Ask questions about your single cell data with natural language
An end-to-end single cell analysis workflow for bench scientists. Construct AnnData objects from S3 + Benchling. Automatic cell type annotation. Query the immunology literature.
Single cell sequencing is one of biotech’s most powerful molecular microscopes. It gives scientists a window into the biochemical state of individual cells, towards understanding precise mechanisms of disease and basic biological processes.
We are well past a decade since this technique came online. As the technology continues to mature, and new kits that increase accessibility and throughput hit the market, the proportion of biological data generated from this modality will only increase.
While the graphical software ecosystem for biochemical assays and other sequencing experiments has become quite mature, accessible tools for single cell analysis lags. The workflow requires a tricky combination of interactive steps, large computing resources and some understanding of high dimensional data analysis to draw real biological conclusions.
Bench scientists need better tools to independently explore and ask questions about single cell data. Giving those with the most experimental context, and extensive understanding of the literature, the ability to play with their own data is important for industry scale improvements in drug development productivity.
Latch is developing a scientific plotting framework backed by large computers that allows biologists to interrogate their data with natural language. We have seen many classically trained molecular biologists complete end-to-end single cell analysis workflows on their own. We highlight the concrete steps with clear graphics here:
Bringing in data and metadata from the cloud
Subsampling counts by condition
QC + filtering
Normalization, dimensionality reduction + clustering
Automatic cell typing
Exploring cell types of interest, re-clustering + querying immunology literature
Bringing counts data and experimental metadata from the cloud
Often the biggest blocker to getting started is accessing data. Here scientists can query AWS, GCP, GDrive for counts and Benchling for metadata to construct a AnnData object for downstream analysis.
Subsampling by patients and tissue type
Scientists are interested in slicing the counts object by, eg. individual patients of interest or specific tissue types.
QC and filtering
Here biologists can generate gold standard quality control plots and use the data to visually remove low-quality cells and genes.
Normalization, Dimensionality Reduction, Clustering
The standard high dimensional analysis steps are exposed to biologists to create the intuitive clusters for more interesting questions.
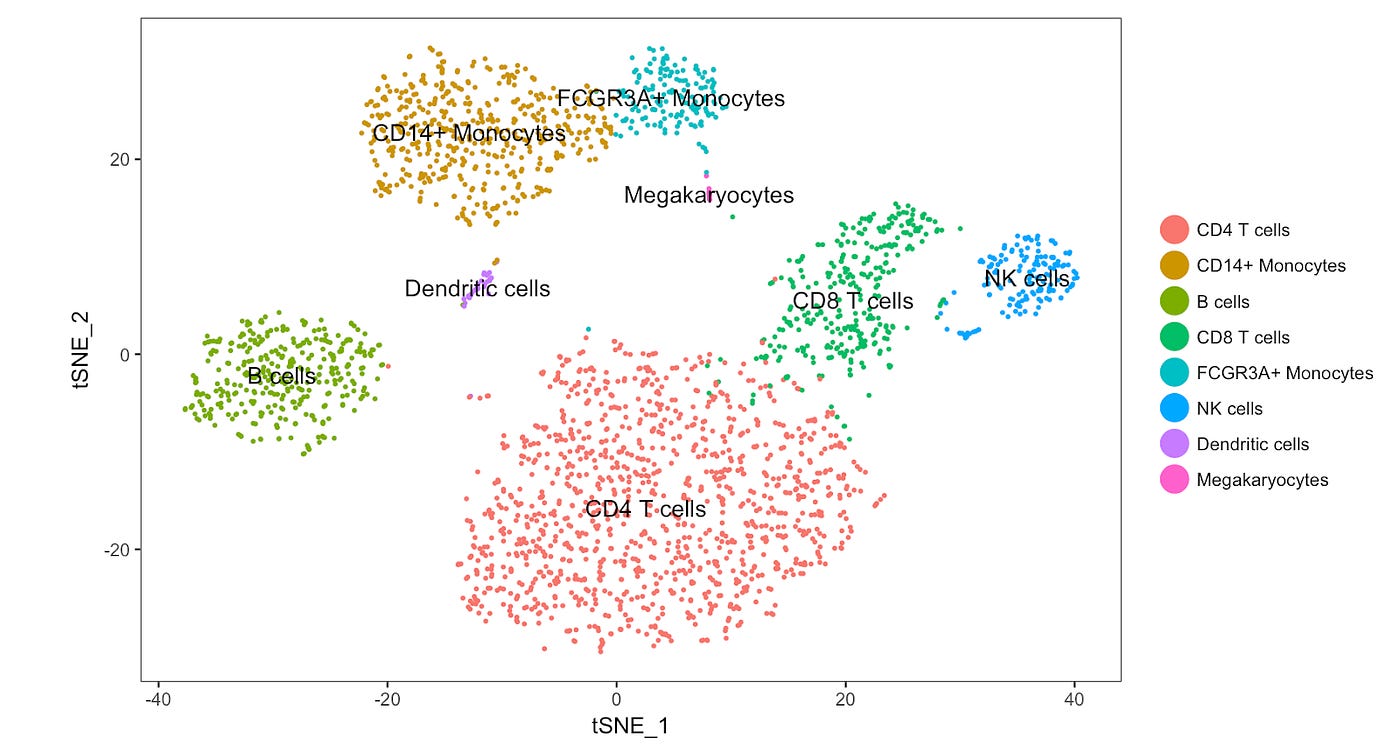
Automatic cell type annotation with machine learning
Machine learning guided cell typing has become quite powerful in the past few months. We expose some of these tools and allow biologists then individually verify and question these annotations with downstream exploration.
Exploring cell types of interest
Scientists can play with the cluster, eg. hovering over interesting cells for contextual metadata, looking at the expression of interesting genes.
Re-clustering to identify fine-grained cell subtypes
Biologists can repeat the standard clustering workflow after focusing on a particular cell type for detailed introspection.
Exploring fine-grained cell subtypes and asking questions about general immunology literature
Biologists can always query the corpus of immunology knowledge available on the Internet to discover eg. marker genes, pathways, interacting cell types relevant to their question at hand.
Giving exploratory tools to biologists increases the pace and efficiency of research
These tools alone are not sufficient. They sometimes produce incorrect results and should be supplemented by computational teams with an understanding of the techniques.
However, allowing scientists to play with their own data will allow for independent hypothesization, new biological questions and potentially new insights with material impact on drug programs. Consider the result of this self-serve exploration when multiplied out across the tens of thousands of capable and educated minds in industry.
Latch is a team of engineers building modular and programmable data infrastructure for biotech R&D. We are rapidly developing new tools in this space and are excited to partner with new biotech teams.