Latch: A Bio Development Framework for Data-Driven Biotechs
The unsustainable state of data infrastructure in biotech, why biodevelopers need their own development framework, an initial design spec, and how to get started with Latch.
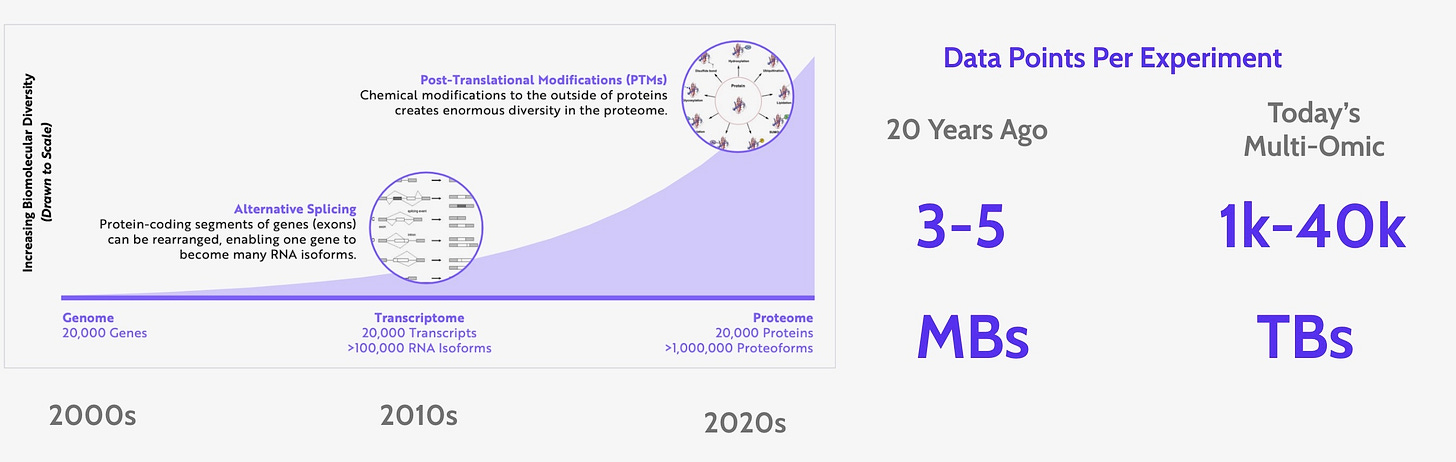
While the growth of biological data over the last ten years has been astounding and has fueled new therapies, drugs, and our fundamental understanding of life, the ever growing amount of data has also become a bottleneck for new discoveries. This new scale of data requires a complex data infrastructure that biodevelopers (bioinformaticians and computational biologists) are spending more and more of their time maintaining, thus spending less time on the data analysis that will lead to new breakthroughs. We propose a Bio Development Framework that makes managing complex biological data infrastructure extremely simple and allows biodevelopers to quickly write, deploy, and monitor their workflows.
We outline:
What is wrong with current biodevelopment tools.
Why biology needs its own development framework.
A Design spec for a Bio Development Framework.
How to get started with the Latch Bio Development Framework.
What is wrong with current biodevelopment tools
Every biology company has data infrastructure, whether they know it or not. In the early days, it seems like a non-problem. You just run a few scripts manually and record the inputs and outputs in Excel or Postgres. But then your organization grows to dozens of people and hundreds of analyses.
As the richness of your experiments grows, your experiment's data gets too big to analyze on your laptop, so you need to set up your cloud environment (e.g., AWS, GCP, K8s) to run your analyses. Then as you start running hundreds of workflows, your company's cloud costs start skyrocketing, and you realize that you need to set up serverless spot instances (e.g., Lambda, AWS Batch) to save cloud costs. And as your organization expands to dozens of people, your biologists ask to be able to run workflows themselves, so you also need to build them intuitive no-code interfaces (e.g., React, Streamlit, RShiny). Then as you start thinking about the regulatory processes required to bring your discoveries to market, you realize that you need to track data lineage automatically (e.g., Docker, Benchling, Postgres, Excel) to make analyzes reproducible down the line. Then as you start to get overwhelmed with analysis requests, you realize that you can automate workflows (e.g., Lambda hooks, Cron) to deliver results faster. And as you hire a few more people to handle this increased workload, you have to start maintaining thoughtful and up-to-date documentation so that new team members can utilize this ever-growing stack.
In an ideal world, you’d use an out-of-the-box no-code platform that contains all the workflows you need and manages all the infrastructure for you (i.e., traceability, cost-effectiveness, ease-of-use). In the real world, this is infeasible, as most of biology, including its complementary computational analysis, is bespoke. New analysis methods are published daily, and biodevelopers need the flexibility to change the underlying code to fit their experiments. From speaking with hundreds of biodevelopers, we learned that no-code platforms lack the flexibility to answer their unique biological questions.
So most of your research needs your in-house biodevelopment team to create and tweak custom analyses, and up to now, that means building custom infrastructure to run those workflows.
What started as simply running a few workflows ends up growing into a very hard-to-maintain in-house system that distracts biodevelopers from data analysis, leading to one of the biggest bottlenecks in biological breakthroughs. The biodevelopers uniquely poised to tackle problems at the intersection of software, data, and biology spend most of their precious time building and managing generic data infrastructure.
But… what if we combine the best of both worlds? A framework that provides the robust out-of-the-box data infrastructure to manage all your workflows and data while also providing an intuitive way to latch into any custom biological code and give it all the same infrastructure.
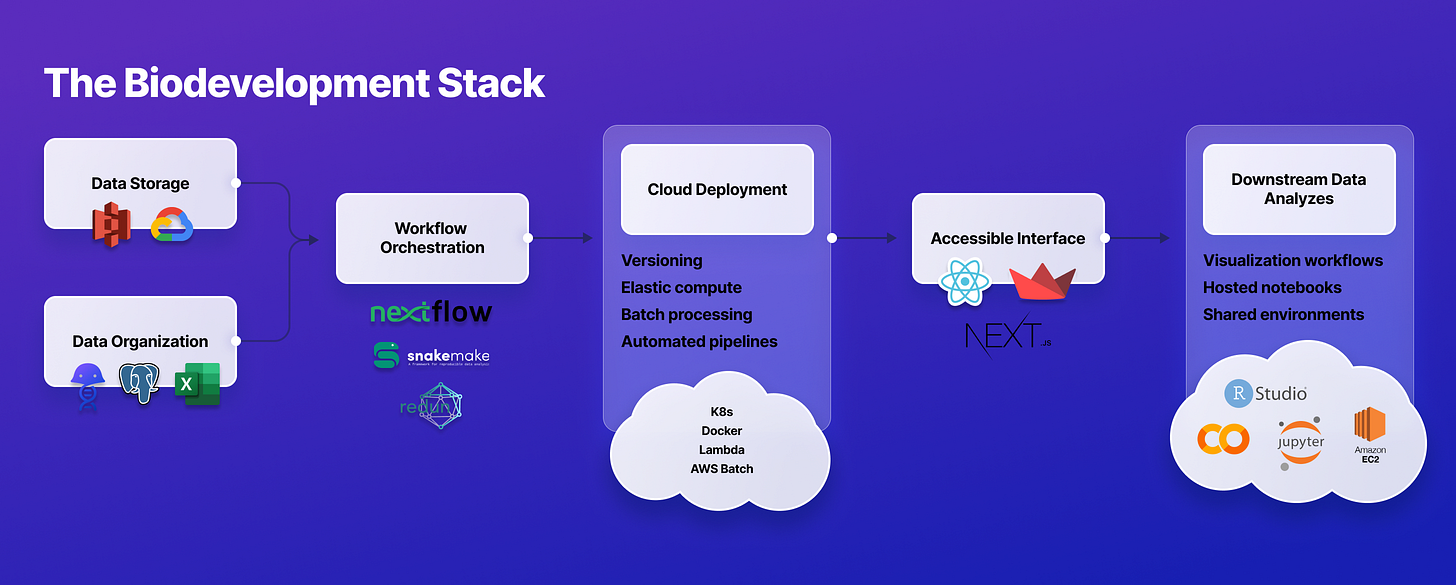
We call that a Bio Development Framework. It makes it much easier for biodevelopers to focus on their analysis workflows while providing all the benefits of managed infrastructure:
Intuitive SDK to upload data and workflows
Simple cloud deployment
End-to-end data provenance
Accessible interfaces
Easy workflow automation
Constantly up-to-date
Active development ecosystem
We believe this framework will empower biodevelopers to spend more time doing their highest leverage work, designing better analyses, analyzing more data, and driving better discoveries.
Why does biology need its own development framework?
Before diving into the framework, it's important to address why biology needs its own development framework. A quick tour through history sketches an answer for that: WebDev and MLops are two great examples of the benefits of specialized development frameworks.
Web development frameworks have empowered software engineers to ship more responsive websites faster. Making a website twenty years ago meant writing raw HTML and CSS, managing the DOM state manually, and tedious reprogramming of dynamic components. Producing and hosting a website used to require a whole team of engineers. Today, developer frameworks such as React, Vue, and NextJS have turned all of that tediousness into managed infrastructure. Now developers can focus on the website's design, architecture, creativeness, and copy.
MLOps (Machine Learning Operations) has empowered engineers to use machine learning in new and exciting ways. Ten years ago, training and deploying a machine learning model meant manually coding layers and optimization functions, rolling your hyperparameter tuning, scaling your models to the cloud, and creating observability tools to monitor distribution drifts in production. Today, developer frameworks such as scikit-learn, PyTorch Lightning, HuggingFace, Tecton, and Ray have turned much of that complexity into managed infrastructure. Developers can now focus on developing better architectures and prototyping new applications.
Biodevelopers deal with similar dynamics as the early adopters of web development and machine learning — lots of time wasted managing generic infrastructure.
Existing development frameworks outside biology (e.g., HuggingFace, React, NextJS) are an inspiration but don't solve the pain points of dealing with big biological data. Biology has unique file types with unique visualizations such as FASTQ, FASTA, BAM, H5AD, PDB, and GTF. "Sample sheets" encoded in CSVs are used to run a batch of workflows. Biodevelopers deal with a combination of bash scripts, C++, R, Python, and Groovy, while trying to parallelize each to hundreds of cores. The concept of automation requires interacting with physical machines from Illumina, Thermo Fisher, Oxford Nanopore, and other bio-specific providers. Data provenance and traceability are more important than in most other industries because of application processes to the FDA. And the visualization tools needed downstream are unique to biology (e.g., Seurat, IGV, ScanPy).
In short, biology needs its own development framework.
Design Specification for A Bio Development Framework
After hundreds of conversations with biodevelopment experts, we distilled the components that stood out as the most foundational pieces of a good biodevelopment framework and will break them down below. These components will naturally evolve as bioinformatics matures as a field. We hope to maintain this design spec as a living document of those requirements.
Let’s dive into them below:
Intuitive SDK to upload data and workflows
Simple cloud deployment
End-to-end data provenance
Accessible interfaces
Easy workflow automation
Constantly up-to-date
Active Development Ecosystem
Intuitive SDK to upload data and workflows
An ideal framework should have an intuitive SDK to wrap all data and workflows in and give them the benefits of the framework. This ensures that biodevelopers can iterate and deploy quickly.
Requirements:
Allows the use of any programming language and workflow orchestrator.
Supports common tools such as Docker and Python.
Github integration to link workflows based on its repository.
Simple Cloud Deployment
It should make running a workflow in the cloud as simple as running it locally. Cloud deployment ensures scalability, security, and accessibility.
Requirements:
A single command for launching workflows on the cloud instead of locally.
Facilitates writing a single workflow and scaling it to hundreds of cores in parallel by simply adjusting a few parameters.
Gives transparent access to logs and run containers for easy debugging.
Uses server-less spot instances with automatic retries to save money on cloud costs. Gives the benefit of a 24/7 super computer but "pay as you go".
Has complete visibility into computing costs. Managing costs on AWS is a known difficult problem.
End-To-End Data Provenance
Reproducibility is key for biological breakthroughs. Every workflow input, output, and code version should be recorded so it can be replicated for the FDA, other regulatory institutions, and colleagues doing similar experiments.
Requirements:
Complete data traceability.
Automatically versioned data and workflows.
A link from every workflow to their respective GitHub repositories and Docker image.
Other metadata tracking such as who launched a workflow, what the inputs were, and when it was launched.
Automatic saving of every parameter and file that goes into and out of execution for replicability guarantees.
An easy interface to query all data, workflows, and executions through SQL or a no-code interface.
Accessible Interfaces (No-code, command line, and API)
A framework should automatically create each interface required for a team to easily perform their job. A developer should easily be able to set up API, and CLI interfaces for themselves, and set up an intuitive UI for their biologists to process and interpret their data.
Requirements:
Automatically created, and intuitive, no-code interface for biologists to use.
Automatically created and intuitive CLI and API interface for other developers to use.
Data visualization capabilities to explore inputs and outputs.
Easy Workflow Automation
Many biological processes are repeatable. Repeatable tasks should be easy to programmatically automate. An ideal framework gives biodevelopers the freedom to automate data analysis to save time on redundant tasks.
Requirements:
Easy automation between data sources.
Automation capabilities so that when data is placed somewhere it automatically runs a workflow on it and then Slacks the results to the scientist who created the sample.
It should track which analyzes have already happened and which have not, then sync everything to the desired state.
Constantly Up-to-Date
A good framework should have a development team that is constantly improving it, adding new features, and staying up to date with the rapidly moving field. As regulatory processes evolve, the framework should evolve with them.
Requirements:
Continuous battle testing against new types of science and different regulatory processes (e.g., Investigational New Drug application, Spatial Transcriptomics).
Rapid improvement based on its learnings.
A contributor team that addresses bugs and issues reports rapidly and fixes them in a timely manner.
Continuously shipping new features.
Active Development Ecosystem
A development tool is only as good as its documentation and community. Clear, concise, and up-to-date documentation coupled with an active community of passionate biodevelopers using the framework is paramount.
Requirements:
Clear and up-to-date documentation for all components.
A quick start guide that makes it easy to get started.
Tutorials and examples for each component.
Community of users who provide guidance, solutions, and examples.
Focus on intuitive developer experience (DX). Wrapping your data and workflows should quickly become second nature.
Getting started with the Latch Bio Development Framework
Two months ago, we open-sourced the SDK to the Latch Bio Development Framework. Biodevelopers can latch their workflows into our infrastructure and get all the benefits outlined above.
Since the release, biodevelopers have built hundreds of workflows, with the most popular of them having thousands of executions each. Everything from protein models running on GPUs like Unirep and AlphaFold2, CRISPR analysis tools with complex dependencies such as CRISPResso and CRISPOR, NGS tools that parallelize to hundreds of cores such as RNAseq and ATACseq, and many more are being added every week. All of them are shared for anyone to use and automatically get all the benefits of the Latch Bio Development Framework.
We have a lively slack community that is quick to answer questions and our team resolves bugs, addresses issues, and ships new features almost every single day.
If you or your team resonated with the value of having a bio development framework to build, iterate, and deploy your workflows you can use the SDK at:
We recommend that you start with a user problem in mind, do the quick start, build a prototype, iterate, and grow from there. And if you do choose to use Latch we would love to help out and hear about your experience (alfredo@latch.bio); we cherish feedback and use it to keep improving rapidly.
We look forward to working with you!
Huge thanks to Nathan Manske for editing my broken English and creating the illustrations. And to Kyle Giffin, Aidan Abdulali, Brian Naughton, Brandon White, Tess Maaike van Stekelenburg, Aditya Joshi, and Blanchard Kenfack for reading and commenting on drafts of the post.