CRISPR Bioinformatics in the Browser
wire #1 // the bottleneck in gene-editing, our Seed round from Lux Capital, and the plan to put the power of bioinformatics in the hands of every CRISPR biologist.
We are living in the age of CRISPR. The years that will be forever remembered as the great inflection point in biology. Those where the scientist picked up molecular scissors to forever change our relationship with our own genetic code — unleashing a renaissance of living medicines; gene and cell therapies made to eliminate global disease and extend the human lifespan.
Quantities of sequence data in CRISPR are growing exponentially — 10x every 2 years. As a result, the infrastructure, computing, and informatics tools needed to wrangle data have caused massive bottlenecks for both labs and new companies to progress. Most biologists don't possess or wish to learn the ever-changing body of best practices needed to orchestrate data across cloud infrastructure, making it difficult for them to leverage the bioinformatics tools required for modern gene editing.
We discovered this problem during our time at UC Berkeley. Talking to over 300 researchers taught us a lot, but one insight eclipsed the rest: talented bioinformaticians are so scarce that biologists often wait weeks for a couple hours of standard data analysis. This bottleneck slows down the whole research cycle, delaying the discovery of precious new therapeutics.
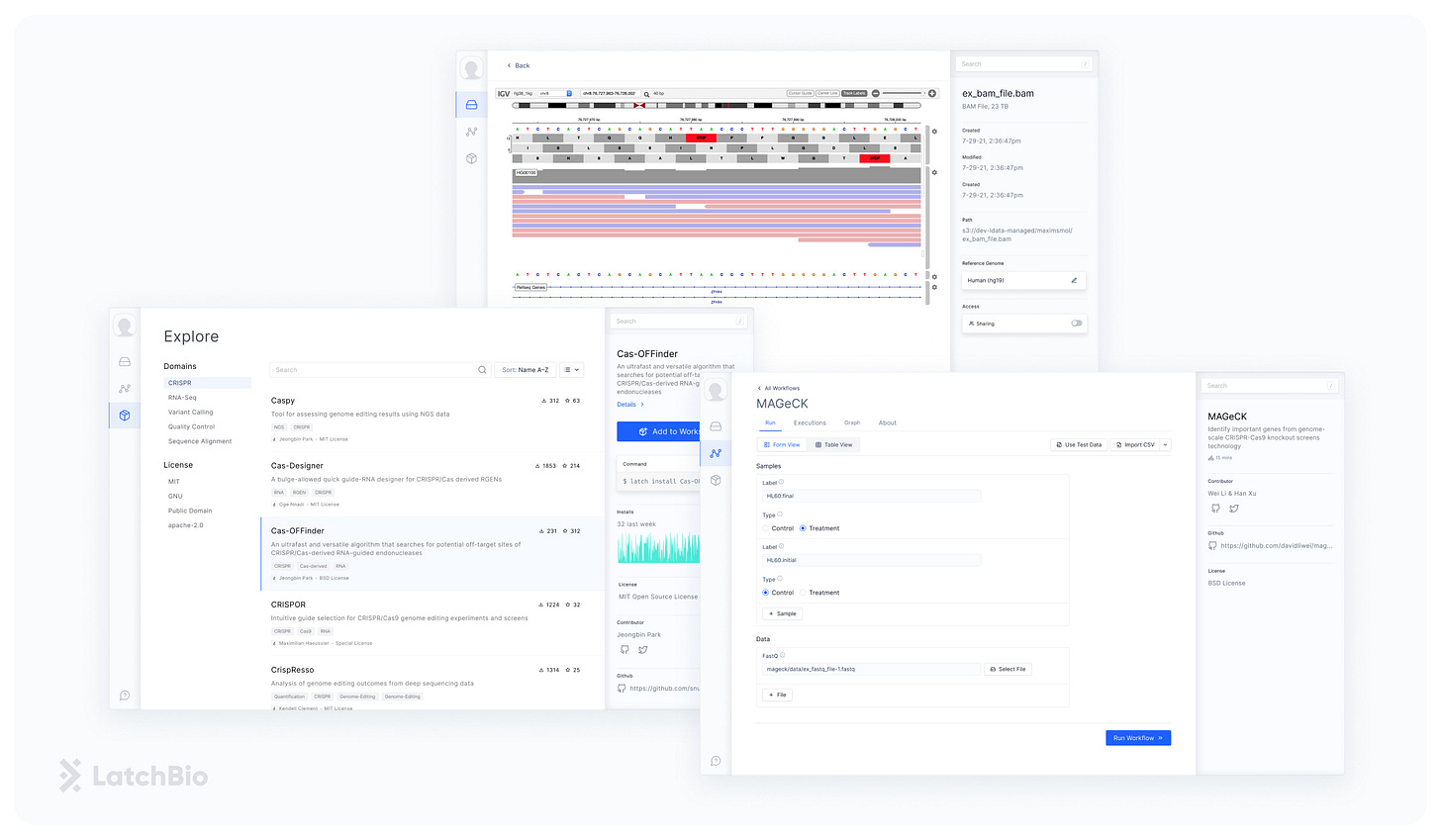
We realized that the bottleneck is deceiving. With the right tools, biologists could run most of these analyses, in minutes, on their own. This is why we built LatchBio, a web platform for biologists to run CRISPR bioinformatics without code. Our goal is to empower the biologist to do their own analysis, massively accelerating their research and freeing up bioinformaticians to focus on developing new techniques.
The LatchBio web platform lets biologists run CRISPR computational analysis without code and it follows a simple loop: upload data, find a workflow, run a workflow on your data, and visualize the results. The cloud-native data store lets biologists bring in data from their computers, Illumina BaseSpace, Benchling, and Amazon’s S3. The workflow explore lets them browse and run dozens of pre-loaded workflows (CRISPResso, Mageck, etc..), that were previously scattered across the web. Finally, the built-in visualizations allow for navigating sequence data effortlessly by running and launching PDFs, Genomic Browsers, or Quality Controls right in the browser.
Pawel Bialk , M.S. biologist at Christiana Care, used to wait for weeks to get back the results of his gene-editing outcomes. Now, he does everything himself in a couple minutes. “The Latch platform will provide immediate access to a wide range of popular CRISPR workflows that will further accelerate our discovery and translational research programs.”
Latch was built with a number of gene editing startups and labs around the world. As of today, our wait-list is open. Every new user will get 100GB of free storage and $100 dollars of free computing.
A core value of Latch Bio is to always be free and open access for academia. In the future we hope to partner with the top scientific journals to make all bioinformatics research reproducible in a few clicks.
We recently raised a $5M seed round from Lux Capital, with participation from Fifty Years, General Catalyst, Haystack, Asimov co-founder and CEO, Alec Nielsen Ph.D, Hexagon Bio co-founder and Head of Data, Brian Naughton, and other key players in the space. We will leverage this funding to put the power of bioinformatics in the hands of every CRISPR biologist.
Latch is a rare opportunity to build a valuable company and accelerate scientific progress at the same time. Being the best CRISPR bioinformatics platform is only the beginning; these problems are prevalent across all of biotech. We envision building one of the great software companies of the 21st century, laying the bricks for the biocomputing revolution.
To experience the Latch platform, please visit www.latch.bio.
P.S. We are hiring.