Generate GUIs and Deploy Bioinformatics Workflows from Python - announcing Latch SDK V2.0.0
The open-source toolchain to solve software in biology: dynamically compile type safe UIs from raw python, serverless pipeline execution, and automatic containerization and versioning
Building software tools for biologists is hard. Developers of these tools often lack the resources and time needed to release their software with graphical interfaces and containerized builds. This results in a fragmented ecosystem of tools that are often unaccessible or hard to use for bench scientists.
We want to make it easier for resource and time-constrained biodevelopers to rapidly add user-friendly interfaces, versioned, reproducible releases to their tools while being able to easily provision resource requirements. That is why over the past few months, we’ve been adding significant improvements to the LatchBio SDK, a framework to create new bioinformatics workflows, provision cloud infrastructure, and automatically generate frontend interfaces from a handful of python functions.
We’ve used this updated framework to build Bulk RNA-seq. This framework has already enabled partners like XCell Bio, Doloromics, and Metcela to turn terabytes of samples into scientific insights.
And with the latest release, SDK users can develop their own production-grade pipelines even faster and with richer functionality. We want to share three key additions that facilitate better local development and publication of large pipelines:
Adding map tasks to parallelize running the same task on a series of inputs, cutting workflow runtimes by several times.
Adding subworkflows and workflow references to reuse workflows within larger pipelines, enabling better versioning and reproducibility.
Enabling easier local development with:
latch preview to hot-reload and view the front-end interface for your workflow
latch exec to get a terminal instance within running tasks on the platform
Latch Terminal-based UI to view all workflows, executions, and logs on the Latch Platform in your terminal.
Let’s briefly take a peek at each and how easily you can use them to develop complex pipelines on Latch:
Parallelize a task across a series of inputs with map tasks
A “map task” lets you run a pod task or a regular task across a list of inputs within a single workflow node. This means you can run thousands of instances of the task without creating a node for every instance, providing valuable performance gains.
We have seen developers use map tasks when:
Several inputs must run through the same code logic
Multiple data batches need to be processed in parallel
Hyperparameter optimization is required
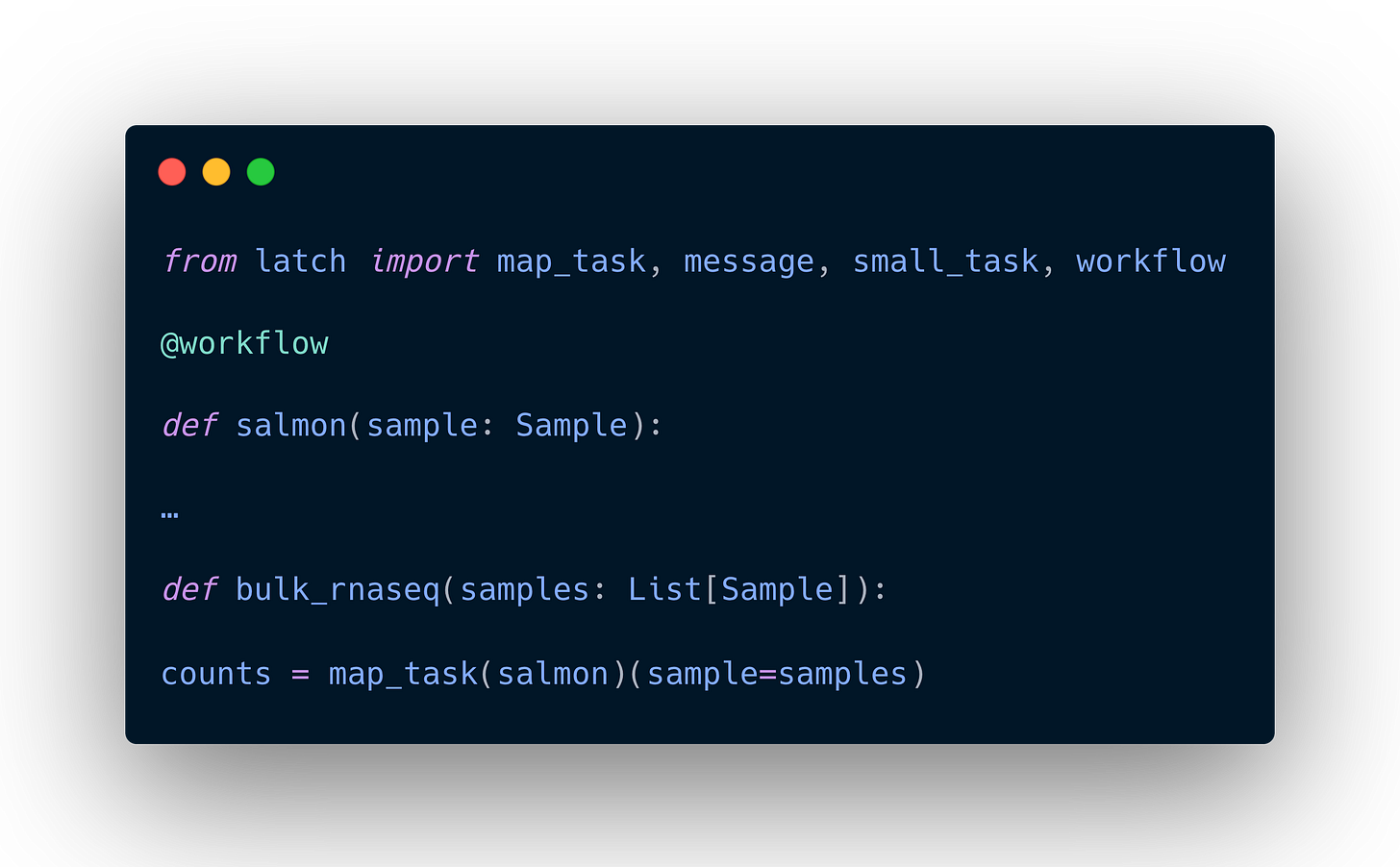
To map a task across a list of inputs, call the map_task on the task function to receive a new “mapped task” function which now accepts a list of inputs as a parameter.
For example:
See a real-world example of how the Latch engineering team uses map_task for our Verified Bulk RNA-seq workflow here.
Reusing workflows within a larger pipeline with Latch subworkflows and workflow references
Subworkflows
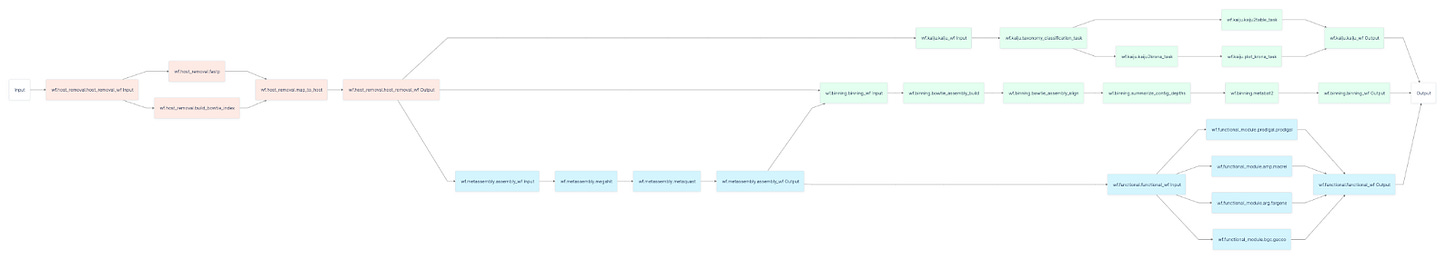
Bioinformatics workflows often contain tens to hundreds of tasks, where each task performs an analysis on different inputs. Subworkflows and workflow references allow for arbitrary composition of workflows within each other, enabling great organizational flexibility as well as reducing code duplication.
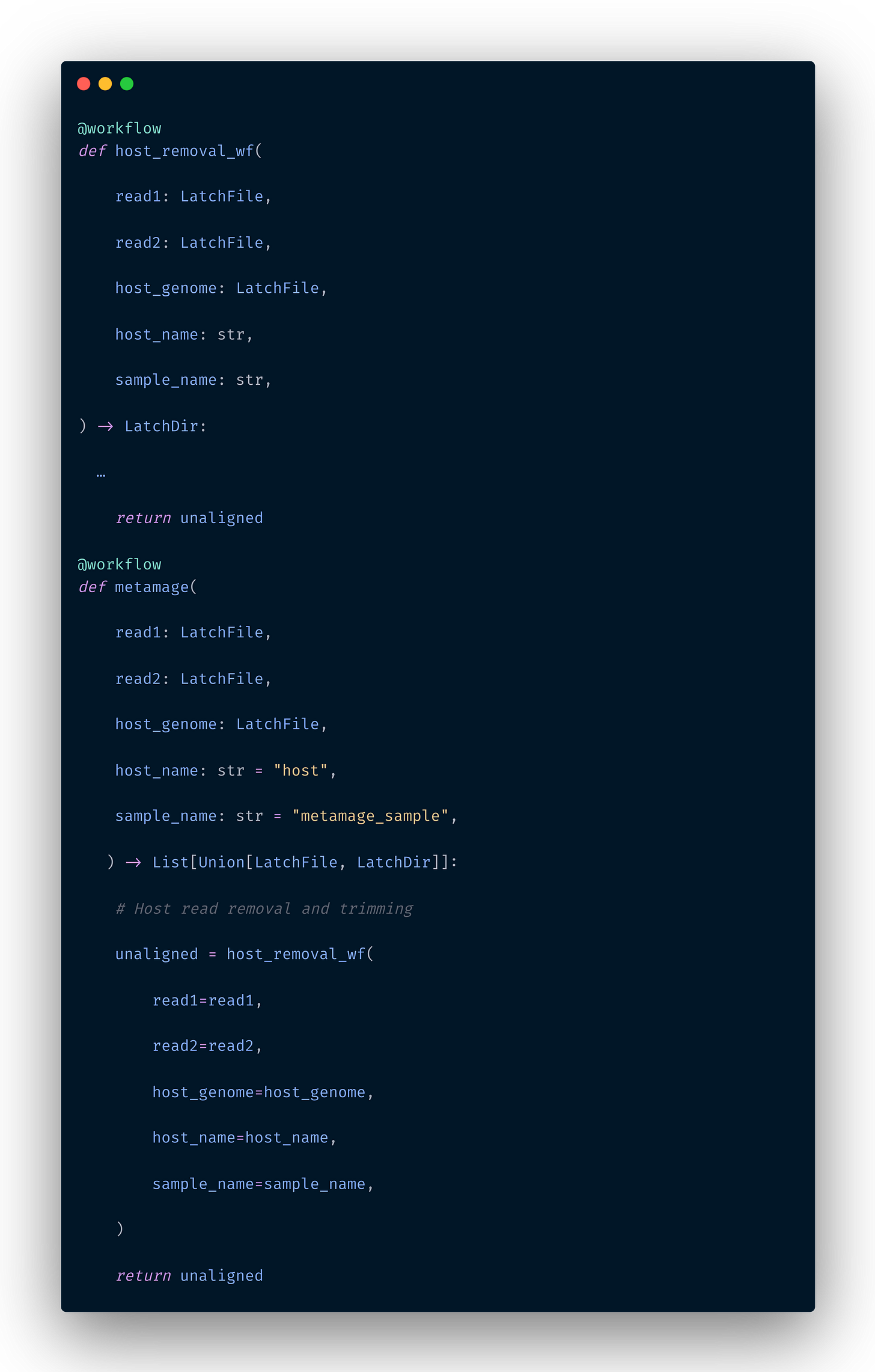
To create a subworkflow, you create two functions with the @workflow decorator and call one inside the other, as below:
See a fully working example of metamage, a taxonomic classification with more than 20 tasks that leverages subworkflows, developed by our Biocomputing Ambassador João Vitor Cavalcante here.
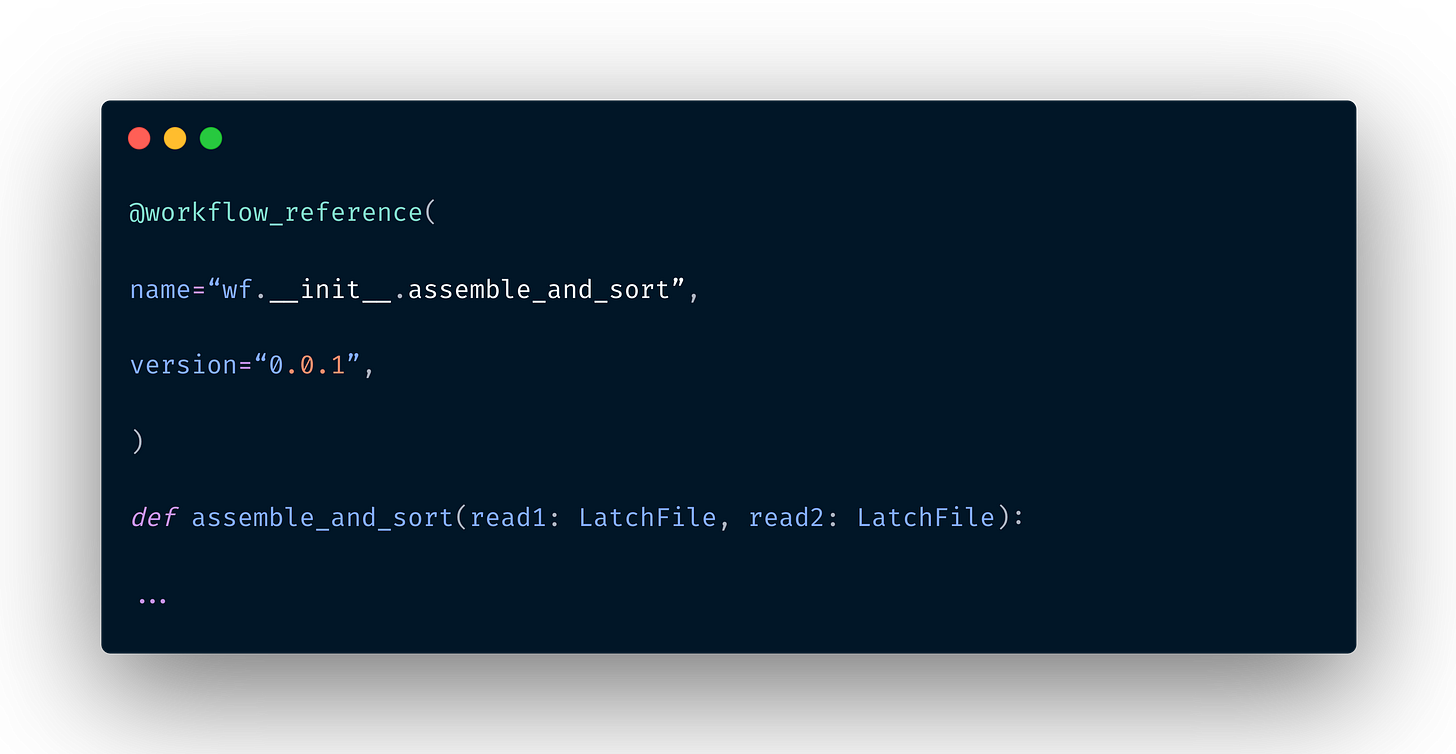
Workflow References
A workflow reference is distinct from a subworkflow in that a workflow reference is a reference to an existing workflow, meaning that entire workflows are reusable in other workflows without duplicating code. To create a workflow reference, simply annotate an empty function with the @workflow_reference decorator as below.
Better Local Development
To support SDK users in developing more complex workflows, we introduced a series of improvements to accelerate local development.
“latch preview” to hot-reload the the user interface of your workflow
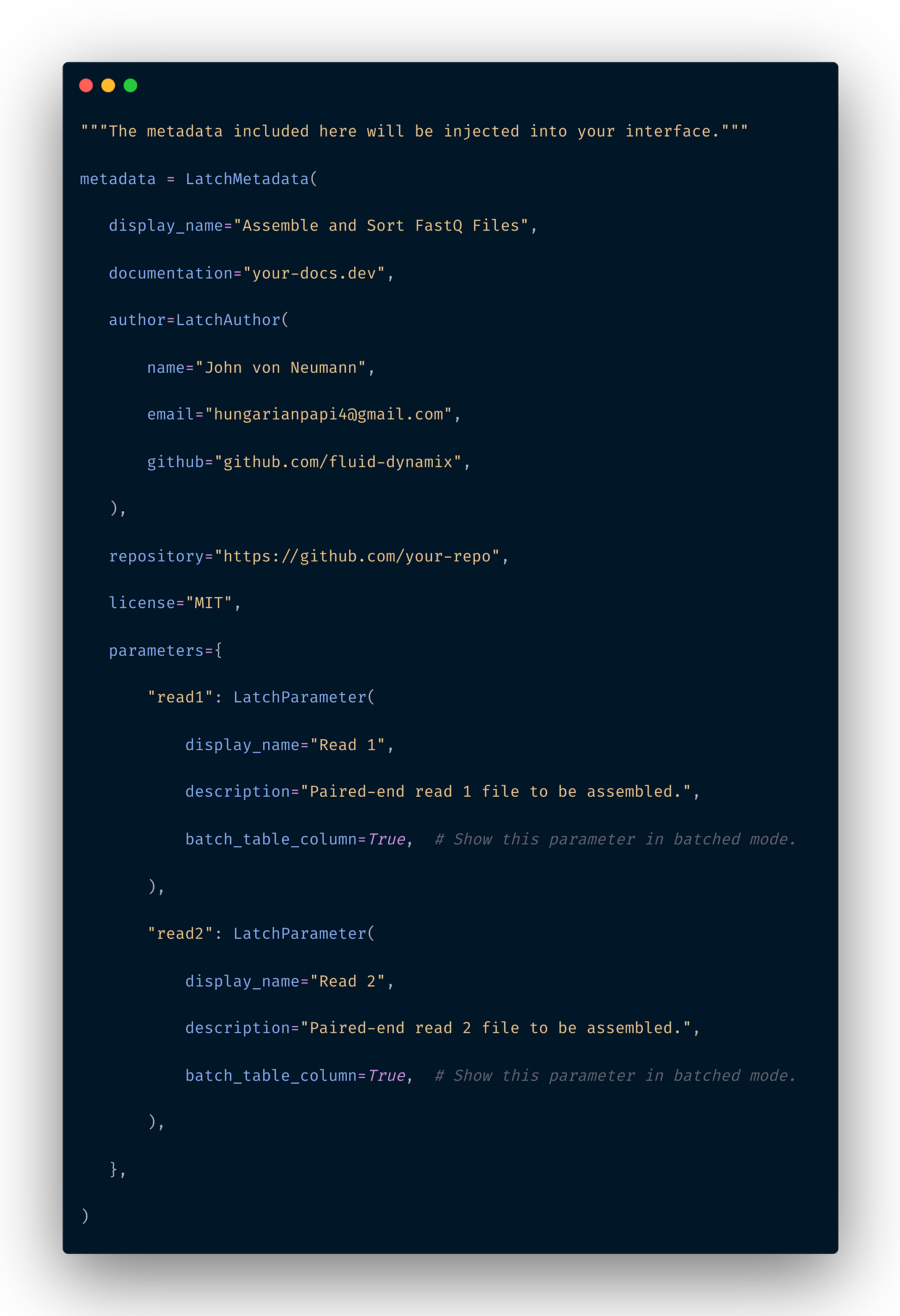
With the latest version of Latch, you can define the user interface of your workflow using Python objects.
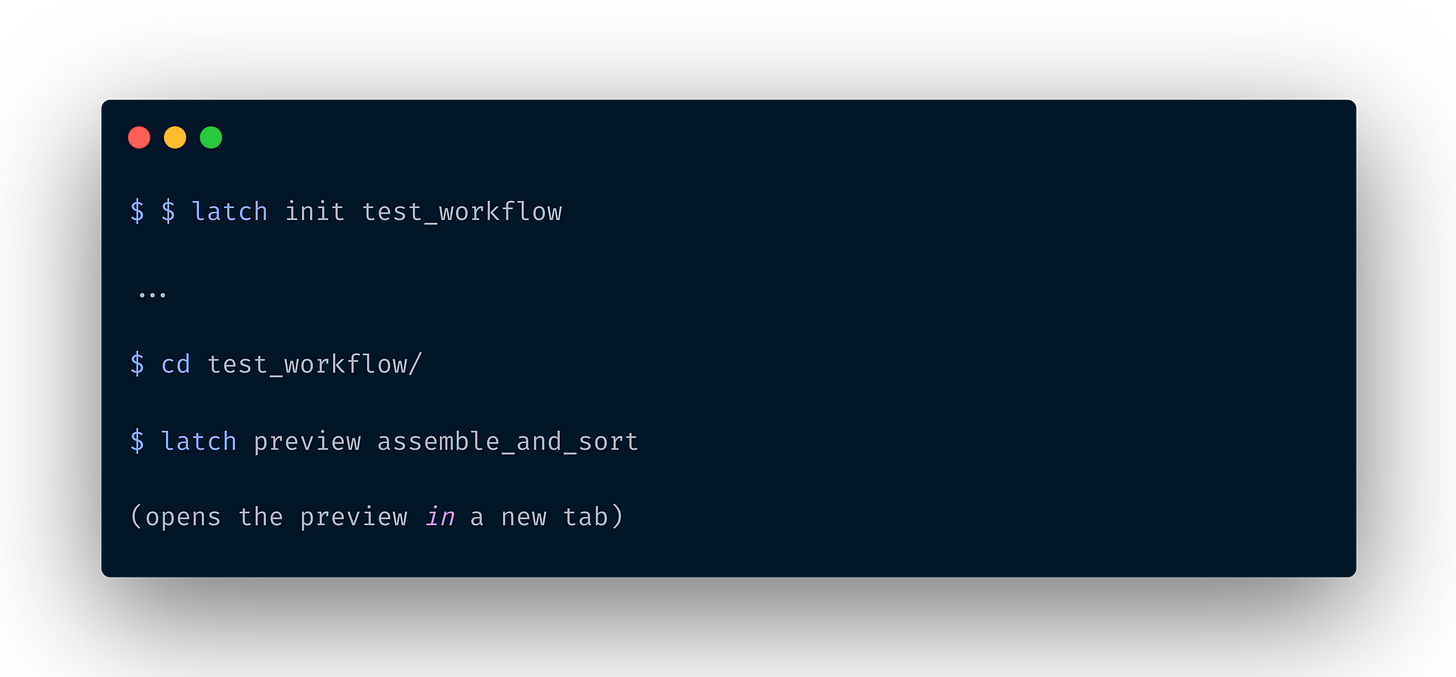
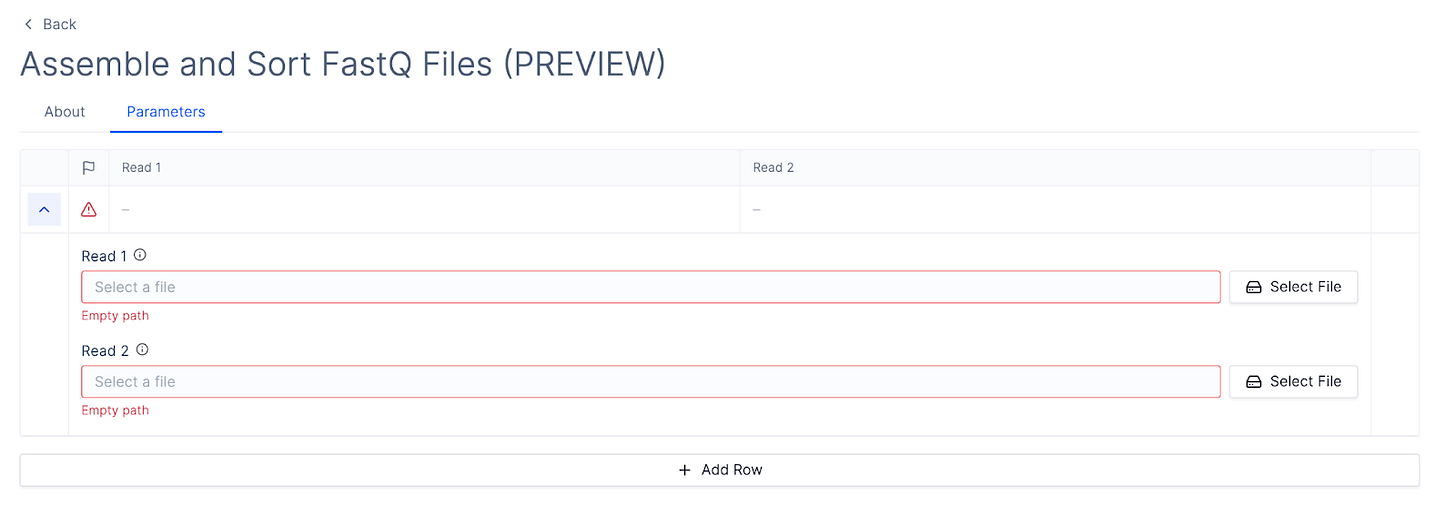
The latch preview command allows you to see a preview of their parameter interface without going through the hassle of registering, building a docker image, and waiting for an indeterminate amount of time between iterations.
To use it, simply navigate to the top-level workflow directory, and run latch preview <wf-function-name>.
This will open up a new tab in the browser with your interface.
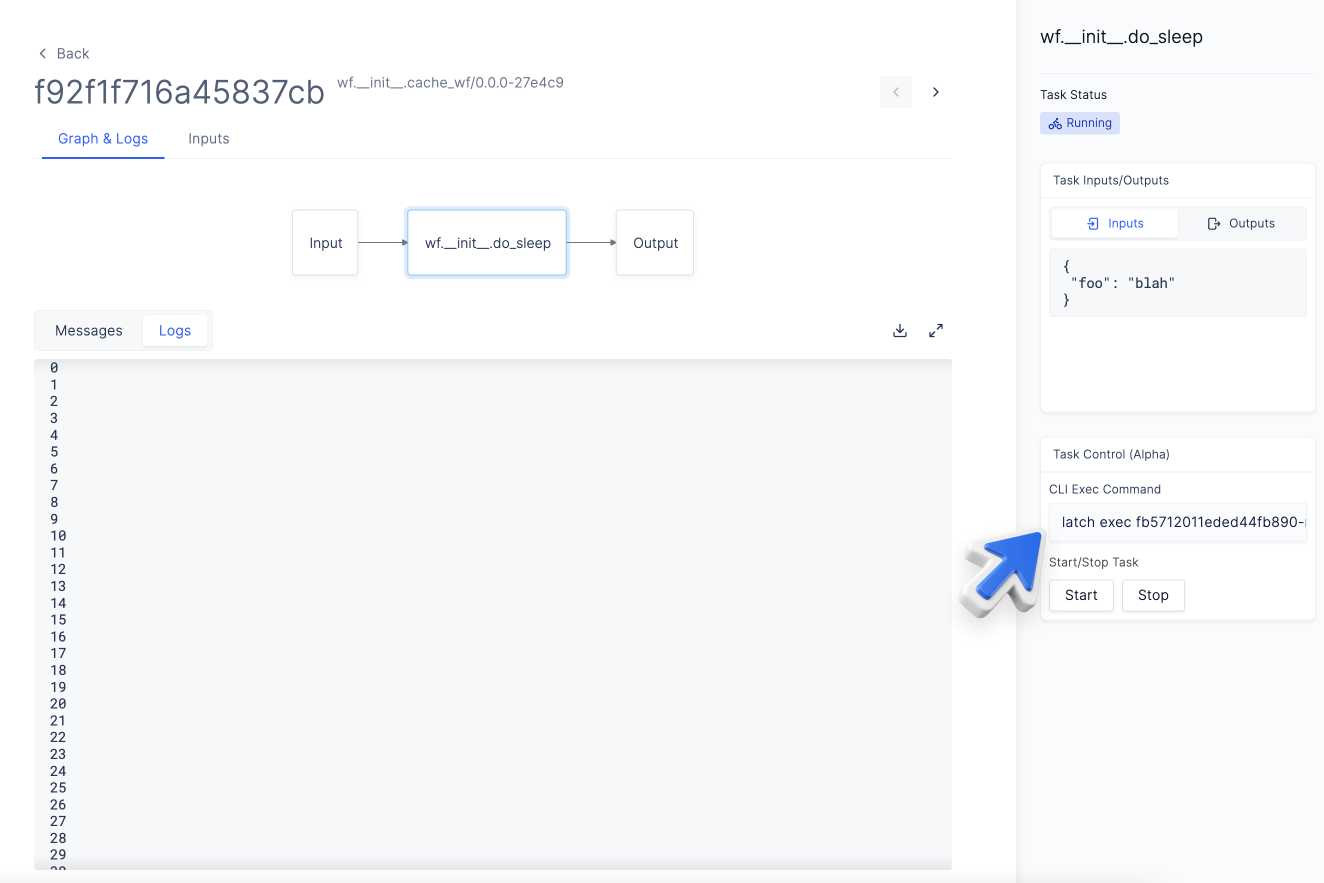
“latch exec” to enter a running instance of a task in the cloud
A crucial component of developing a workflow in the cloud is to be able to debug and understand why the workflow fails in the exact environment that it failed in.
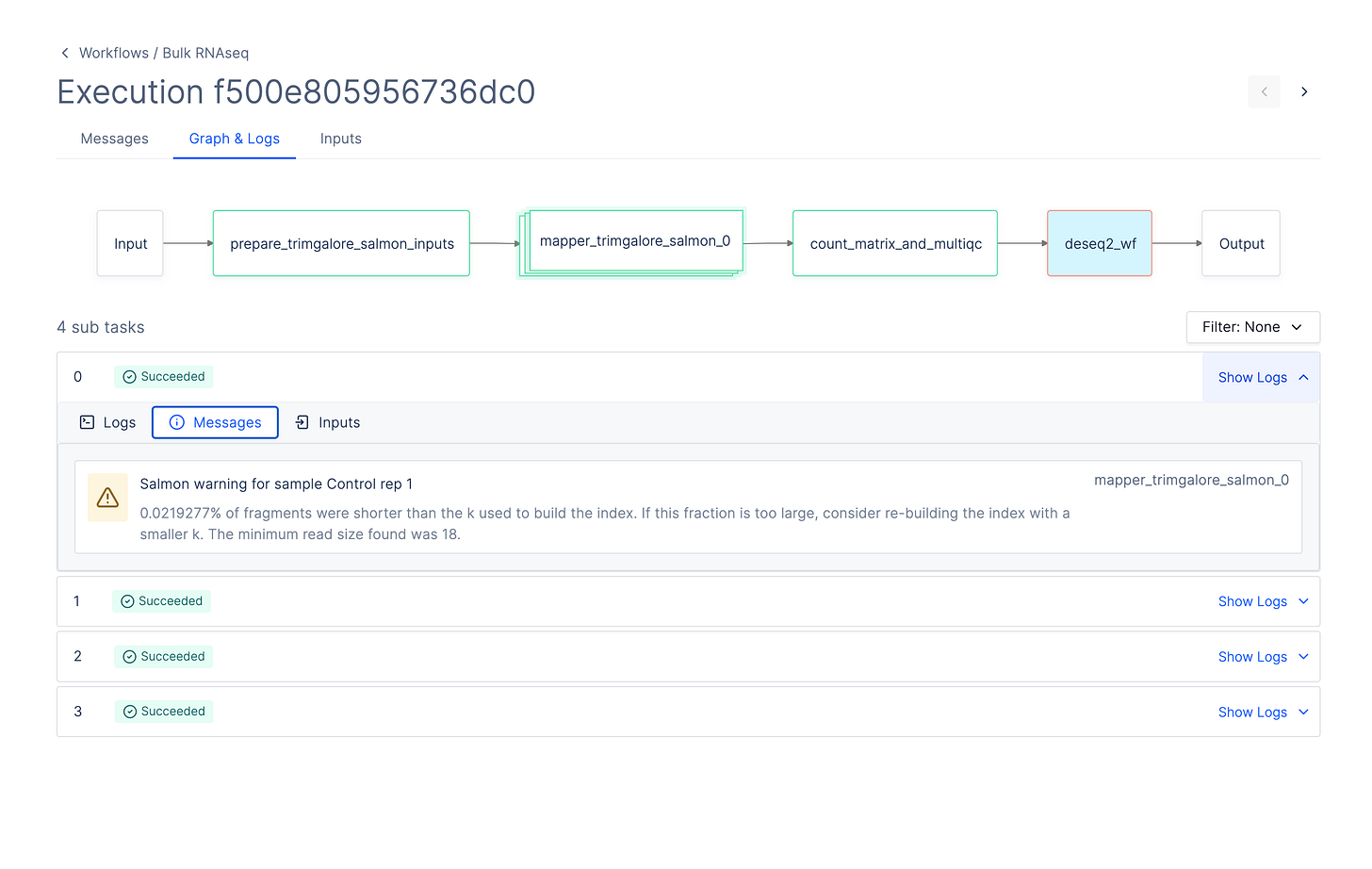
Latch exec allows you to get a terminal instance within running tasks on the platform. To use latch exec, select the task in the DAG of your workflow and copy the latch exec command on the sidebar for easy use.
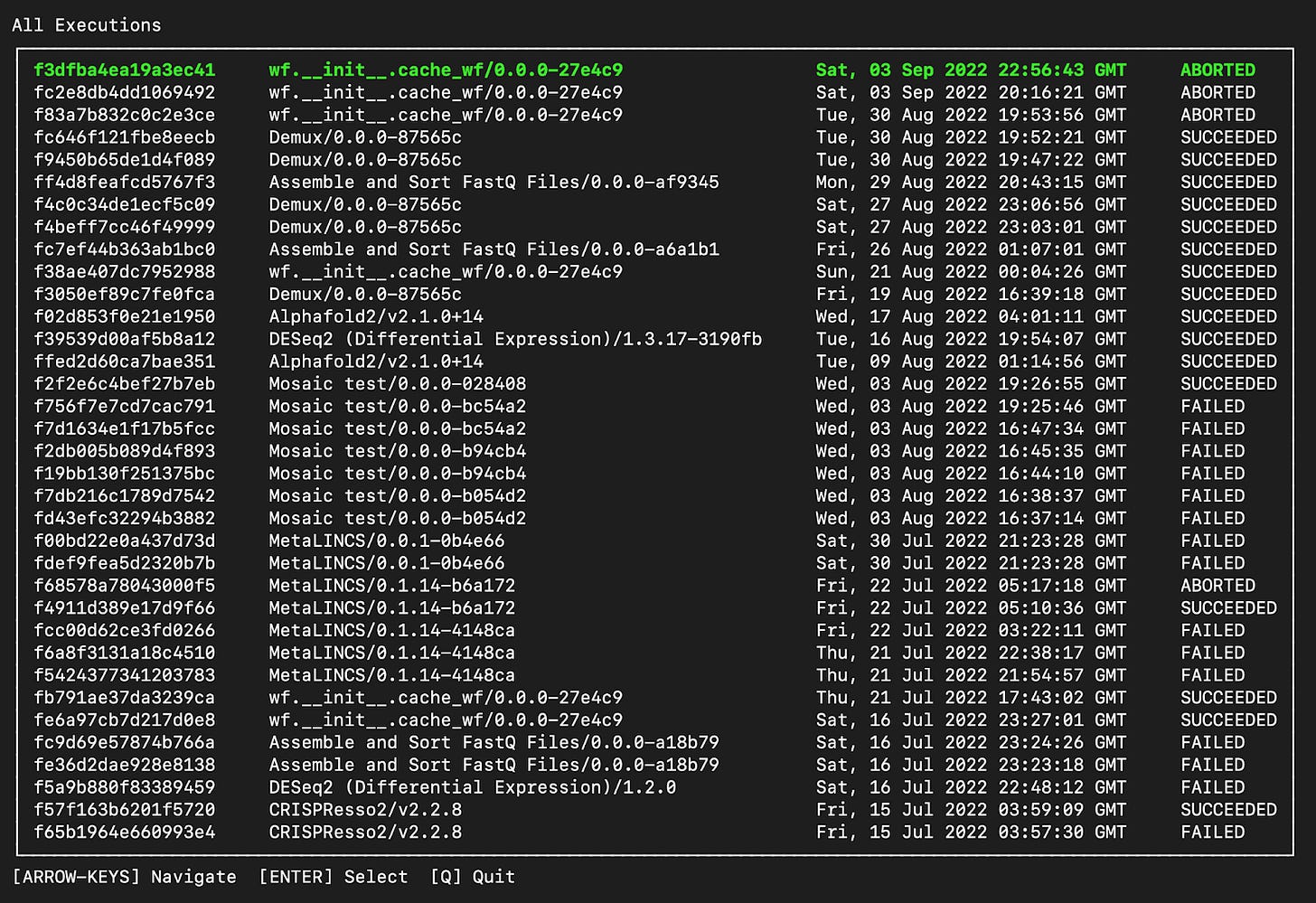
Latch Terminal UI: Develop cloud workflows without ever leaving your terminal
It is often time consuming to have to switch between your terminal and the Latch platform to view executions, workflows, and logs.
We introduced a terminal UI version of the Latch Executions page, allowing you to easily browse between executions, tasks, and logs for all workflows.
To view all executions, type latch get-executions in your terminal:
Now, you can have full observability into your workflows and executions without having to ever leaving the terminal.
What’s Next?
Getting started with the Latch SDK is easy.
Visit our Quickstart and Authoring your Own Workflow to understand how to get up and running with your first workflow in 60 seconds.
Browse Workflow Examples built by our community members and Tutorials to further understand Latch concepts.
Credits
We want to extend our gratitude and acknowledgement to all Latch community contributors @ayushkamat, @kennyworkman, @maximsmol, @r614, @rohankan, @mrland99, @nahid18, @jvfe, @JLSteenwyk for helping us achieve this milestone.
If you want to be part of the community, join us in our Latch SDK Slack.
We’re excited to have you join the biocomputing revolution and start building scalable, reproducible workflows with the Latch SDK.