Introducing Pollock: a single-cell browser for dynamic transformations on millions of cells.
Browse, edit, or create annotations, sub-cluster and re-do dimensionality reduction, visualize differential gene expression, track all transformations and analyses, and much more...
Single-cell datasets are large and unwieldy. Current cell browsers are static and don’t allow for the computationally intensive transformations needed to truly explore their complexities, such as zooming in on a specific cell cluster to find more granular cell sub-types or doing differential expression between multiple hand-labeled cell clusters.
We wanted to make a single-cell browser that had the power to do all of this while remaining extremely easy for anyone to use.
Using our visualizer, we replicated a paper by Olah et. al and rediscovered a unique gene network in Alzheimer’s.
If you know any scientists who would take advantage of a dynamic, interactive, and fully traceable single-cell visualizer, share it with them ;)
Easily browse, edit, and create annotations
Browse existing annotations, manually edit the annotations, run Louvain or Leiden to generate new annotations, or use machine learning to create cell-type annotations.
Subcluster and re-do dimensionality reduction
Powered by a supercomputer, Pollock enables scientists to do live transformations. Zoom into a cell population by sub-clustering. Then re-run PCA, UMAP, or TSNE on that sub-cluster to explore new insights.
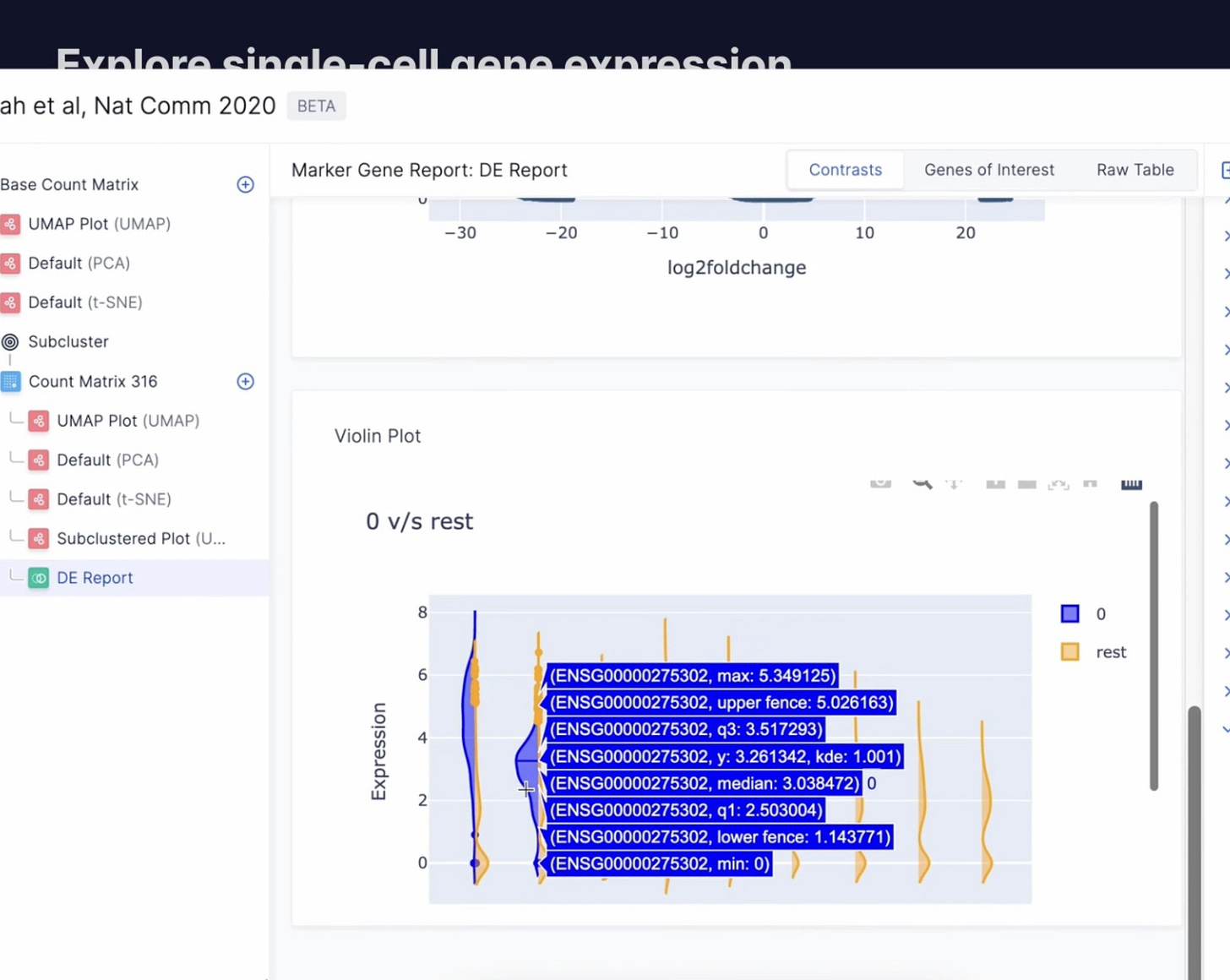
Visualize differential gene expression
Explore differential expression across any two clusters of cells with intuitive visualizations. Or visualize gene expression heatmaps directly on a UMAP.
Track all transformations and analyses
Create a new count matrix every time you subcluster. Track the inputs and outputs of every transformation.
Resources
Check out all the capabilities through this great tutorial by @lehannahle: https://latch.wiki/getting-started-with-pollock
Follow along as @AldermanEva uses Pollock to replicate an Alzheimer's publication: https://latch.wiki/discovering-microglial-subclusters
Try it now at: http://console.latch.bio (it is pre-loaded with test data).
As always, please send us all your questions, comments, or feedback. 🙏🏼