LatchBio releases an integration for Nextflow
drop-in existing workflows // monitor + optimize resources // Github integrations // explore the execution graph // read from graphical samplesheets // build dashboards to share results
Towards building a long-lasting and widely adopted data infrastructure for the biotech industry, LatchBio is excited to release a native Nextflow integration.
Over the past decade, computational scientists have consolidated around Nextflow to build, scale and share scientific workflows. A vibrant community has emerged around the framework to share best-practices and maintain public libraries of gold-standard tools.
While our team initially had technical reservations about supporting Nextflow, we listened to the community and realized we misunderstood the value of the framework. This language, and its supporters, are the foundation for a new era of global and open science, where long lasting projects outlive single PhDs and labs pool their knowledge from all corners of the world.
After building closely with incredible biotechs for nearly a year, we are releasing new tools to develop and host Nextflow workflows in an end-to-end data infrastructure . Our team pledges our full support to the community and is excited to refine and extend this integration for years to come.
We’ll highlight:
The developer experience and the exact steps to drop-in existing projects
New tools to develop and interact with workflows, like resource dashboards, graphical DAGs and Github / container repository plug-ins
A library of hosted nf-core workflows
Integrations into a full analysis lifecycle - pulling from metadata schemas, writing exploratory code in Jupyter / RStudio + building dashboards with workflow results
Host existing Nextflow workflows with minimal modifications
Developers can drop-in existing Nextflow projects and configure their workflow in 3 steps:
Generate a single Python file to configure your interface from Nextflow code
Modify or extend this file
Register the workflow with Latch
Latch preserves the portability of Nextflow code and prevents disruption to development flows with platform-specific modifications.
New Tools to Develop and Execute Nextflow
Self-serve interfaces for scientists with Python
Exposing routine bioinformatics workflows to scientific teams for self-serve execution is challenging. Dozens of parameters, with complex and error-prone values, need to be understood and filled out by experimentalists.
Use Python, in a single standalone file, to organize interfaces into sections and component hierarchies, ingest complex values like sample conditions and give parameters graphical context, like hover-over tooltips, metadata and alternative display names.
Visualize the workflow graph
Navigating a workflow graph allows wet-lab and computational scientists alike to reason about the data flow, monitor execution progress and debug errors.
Debug workflows by relaunching from failed tasks
Modify code with a print statement or a quick fix and relaunch from the failed task. Avoid filling out new parameter values by hand or waiting for upstream sections of the workflow to re-execute.
Inspect and optimize resource consumption across tasks
Clearly see per-task resource utilization to cut cost and increase workflow performance.
Associate workflow versions with Git commits
Recover a snapshot of your workflow code for specific executions.
Integrate with private container repositories
Use existing container images with secure integrations to private repositories.
Run gold-standard community workflows with nf-core
nf-core is a community-curated collection of gold standard bioinformatics tools for popular assays. Biotech teams can access the nf-core library, and take advantage of graphical interfaces and scalability, on Latch.
Integrate workflows in a broader ecosystem of data analysis tools
Bioinformatics workflows are a small component of an end-to-end analysis lifecycle. Hosted Nextflow workflows are often linked with other modular components of the Latch platform.
Read and write from shared metadata tables
Nextflow workflows can read and write from shared metadata tables, often in the form of samplesheets, that capture experimental context from the wet lab.
Explore workflow results in hosted RStudio and Jupyter environments
Pull outputs into environments for exploratory data science from a shared filesystem.
Build interactive dashboards to communicate results to scientists and leadership
Construct reactive and programmable plot layouts to share results.
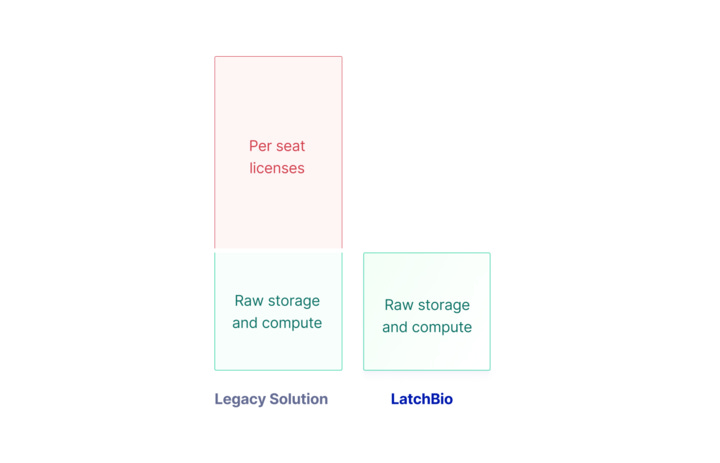
A business model that encourages your entire team to access and understand your data
Onboard your entire company to run workflows and access results.
Usage-based pricing with no seat-based licenses or fees. Pay as you go and only for what you use.
Host Nextflow for your Team
Beyond hosted Nextflow, Latch is a modular and highly programmable data infrastructure designed to orchestrate wet + dry lab teams for deeper + faster biological consensus. For more information about the components of the platform, check out our documentation:
We work with over 100 biotechs, ranging from fledgling startup to top 20 biopharma, and invest heavily in support and customer obsession. Our bioinformatics and engineering services team would love to partner with you on your next project.