Launching 1,000s of Bulk RNA-seq with Latch Verified
A robust, open-source pipeline built with Latch SDK
Today, we’re excited to announce the launch of Latch Verified Bulk RNA-seq - a curated, end-to-end RNA-seq pipeline that takes in FastQ files and converts them to count matrices, differentially expressed genes, and top enriched pathways.
For any area of bioinformatics, the set of possible tools and algorithms is vast and constantly evolving. As a biotech company grows and onboards new assays, it can take weeks, if not months, to build new bioinformatics pipelines, provision the necessary infrastructure, and build an intuitive interface that can be exposed to scientists.
By serving as a bioinformatics CRO for our design partners and observing thousands Bulk RNA-seq executions, we identified the three most common reasons why current out-of-the-box pipelines fall short:
1. High barrier to deriving insights
Oftentimes, scientists receive samples from sequencing cores and want to find differentially expressed genes. However, they have to go through multiple, discrete workflows on different platforms before receiving an analysis report. Learning what workflow to use and how to use them properly takes valuable time and expertise that should have been spent interpreting results and conducting experiments.
2. Complexity in Scaling
Bioinformatics is dominated by terabytes of data and workflows that require multiple CPUs or GPUs, making sharing and scaling pipelines difficult. It often takes engineering teams months to build a robust cloud infrastructure necessary to support the ingestion and execution of bioinformatics pipelines.
Off-the-shelf tools don’t perfectly work either, as they often require scientists to design a complex sample sheet before they can launch multiple samples. Such steps may seem trivial with small sample sizes, but if scientists have tens of experiments and hundreds of samples per experiment, the process quickly becomes error prone.
3. Lack of flexibility
Given the diverse tools and unique needs for each team, there is no perfect “one size fits all” solution. Although legacy no-code tools allow scientists to run workflows themselves, it hinders the bioinformatician’s ability to make changes, such as swapping out an alignment & quantification tool or adding a preprocessing step to the workflow. Further, depending on requirements for speed and resources, a bioinformatician might want to fine-tune resources (CPUs, cores, rams) to each step in the pipeline, which is impossible with no-code solutions. The tool and infrastructure lock-in remains a sticking point for teams to fully adopt an off-the-shelf pipeline.
Teams need a 24/7 supercomputer on the cloud that can run RNA-seq analysis on TBs of data, while being intuitive for scientists and flexible for bioinformaticians.
Introducing Latch Verified - A Bulk RNA-seq Workflow built with Latch SDK
Built and open-sourced using the Latch SDK, our bulk RNA-seq makes it easy for bioinformaticians to clone and modify Latch’s existing pipeline, while being highly intuitive for scientists.
Step 1: Bulk RNA-seq to generate count matrices
To run Bulk RNA-seq on TBs of FastQ’s, scientists can simply click on the folder that contains all sequences.
Latch automatically infers sample names and strandedness from folder and file names, and produces a sample sheet that’s guaranteed to work with our RNA-seq pipeline.
Based on the file name, Latch also detects whether the sample is a single-ended or double-ended read, while automatically inferring the sample strandedness.
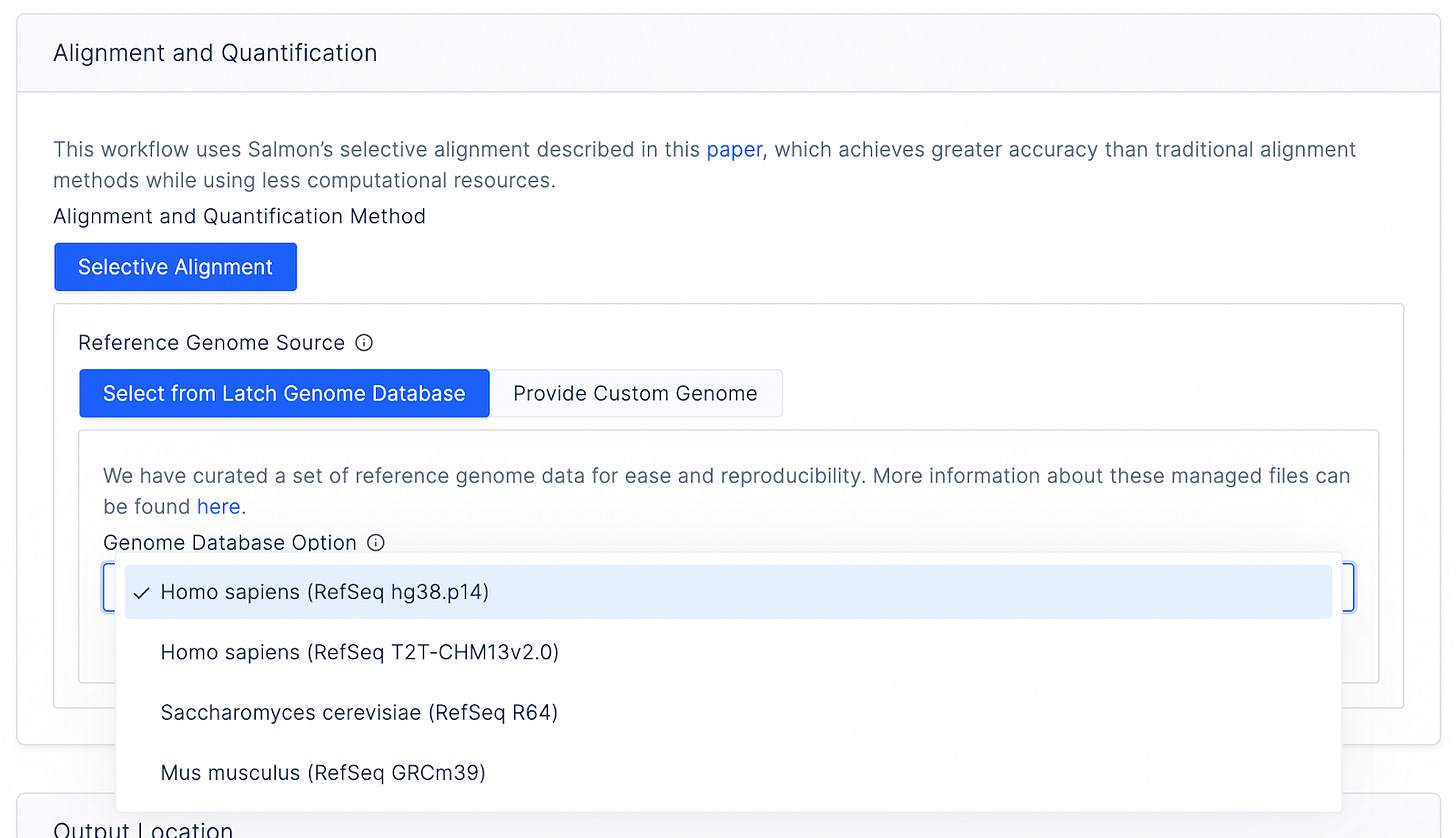
Further, instead of asking scientists to upload their own genome references and annotation files, which can be relatively heavy and hard to find on the Internet, Latch provides a list of curated genomes for humans, yeast, and mice.
By making sample ingestion extremely easy and adding abstraction layers over parameters such as strandedness and reference genomes, our Bulk RNA-seq eliminates some of the most common errors encountered by scientists.
Step 2: Run differential gene analysis to find top differentially expressed genes
Instead of visiting another platform for differential analysis, Latch allows you to directly ingest the count matrix from the previous Bulk RNA-seq workflow, or bring your own count matrices from somewhere else. If you run multiple Bulk RNA-seq in different batches, you can also easily combine count tables.
For the design matrix, scientists can upload a CSV containing sample IDs and conditions. Latch allows you to fine-tune complex experimental design by specifying which column corresponds to an experimental condition versus a confounding variable.
Scientists can have complete visibility into what pairwise comparisons will be performed, as well as the design formula in R.
One of the unique features of our Differential Gene Analysis workflow is that scientists receive a visualization app at the end. Think clicking on a file and it opens up an RShiny or DashApp for interactive, point-and-click querying and plotting.
For example, scientists can easily add genes of interest to have them automatically displayed on heat maps, volcano plots, and MA plots.
See a sample Differential Gene Expression Visualization on Latch here.
Step 3: Enrichment Analysis
Finally, our Enrichment Analysis workflow directly takes in comparison table output from the differentially gene analysis workflow, and returns the top 20 pathways from KEGG, mSigDB, and GO databases.
Once again, scientists can dynamically interact with our visualizations. For example, all genes on the KEGG pathway diagram are clickable – simply select a gene and visualize them on a bar plots for top differentially expressed genes in a specific enriched pathway of interest.
See our sample Pathway Enrichment Analysis report here.
Finally, A Thank You to Our Partners
Latch Verified Bulk RNA-seq wouldn’t be where it is today without the generous support of our design partners and biocomputing ambassadors community.
We want to extend our thanks to:
XCellBio, who is harnessing the power of high-throughput data processing to optimize cell therapy and drug discovery. XCell saves the costs of hiring 1 FTE and 3-6 months setting up infrastructure by running their bulk RNA-seq samples on Latch.
Metcela, who is revolutionizing the way we treat heart failures. By collaborating together, Metcela was able to find top differentially expressed genes and pathways for fibroblasts.
The Love Lab, who is working towards the personalization and actualization of medicine. Latch allows all lab members to utilize RNA-Seq for their projects without running scripts on the local cluster, and provides central locations for all RNA-Seq datasets.
Amit Halkhoree, Thyago Leal, and David Mai for robust weekly testing and feedback on the pipeline.
With verified workflows, Latch aims to create an intuitive and battle-tested pipeline that scientists can rely on to derive insights for their next experiments.
To get started, we have prepared datasets for two academic papers, so you can replicate their results on Latch. Running Bulk RNA-seq, Differential Gene Analysis, and Pathway Analysis through them is extremely easy: Simply head to each workflow linked, select “Test Data” to choose your paper of interest, and click launch.
We are trying to compare transcriptional reprogramming of prostate cancer cell lines under different environmental conditions that mimic tumor microenvironment. The biggest challenge we faced is slow speed in server set-up, data migration, pipeline execution, result visualization.
Latch Bio offers a "One-Stop-Shop" solution to streamline all the above mentioned steps, without the hassle of setting up our own infrastructure. — Xueyang Feng, Senior Director of Computational Biology, XCellBio
This is just the beginning. To learn more about Latch Verified:
Visit our GitHub repository to see source code for bulk RNA-seq, differential gene expression, and pathway enrichment analysis.
Try it out at Latch Console and dive into our End-to-End RNA-seq Analysis Tutorial
We are excited to be with you on your RNA-sequencing journey!