The Open Source Stack for Biological Imaging
A speedrun through scientific imaging tools, what each of them do, concrete scientific use cases and infrastructure principles.
Imaging allows scientists to actually see biological structures and processes in modern research workflows. From a developing zebrafish embryo to monitoring neural activity in two-photon microscopy, these datasets are growing in complexity and size, with increasing resolution and fluorescent channel count.
In this essay, we’ll dive into some open source imaging tools and what they actually do. We’ll walk through Napari, ImageJ, Cellpose, CellProfiler, Suite2p, highlighting the original motivation for each tool and how it fits into common lab workflows. We’ll then construct some real-world scientific use cases, from segmenting microglia in brain slices to high-throughput compound screening in cancer models. Finally, we’ll explore some data infrastructure and software principles that lead to better analysis for scientists.
Beloved imaging tools (and what they do)
Napari
Napari was released in 2019 to annotate, segment and “track” (the stitching of objects between frames of a video) increasingly large multidimensional images. This coincided with the growing number of simultaneous fluorescent channels used in emerging imaging techniques that generated unwieldy, and very big, files.
It leverages GPU-acceleration and a layer-based design + plugin system. Its API is highly readable and easy to understand.
ImageJ
A longstanding, Java-based image analysis tool with a massive plugin ecosystem.
Wayne Rasband initially wrote its predecessor, called “Image”, for a PDP-11 minicomputer with 64KB of memory back in the late 70s. Rasband rewrote the tool several times and eventually settled on Java for the portability benefits of the JVM to create ImageJ. The tool is a bit dated but has a solid array of features for basic processing, along with a rich plugin library.
Cellpose
Developed by Marius Pachitariu and Carsen Stringer at the HHMI around 2019, Cellpose filled a gap for off-the-shelf cell segmentation.
It uses deep learning models trained on multiple cell types, reducing the need for researchers to curate their own specialized datasets. Cellpose’s user-friendly interface and strong performance across varied microscopy images accelerated its popularity, helping labs tackle segmentation challenges with less guesswork.
CellProfiler
CellProfiler was among the first open-source platforms (circa 2005) to tackle modular, repeatable pipelines for high-throughput image analysis.
By providing a graphical interface and a modular pipeline system, it allows teams to measure hundreds of morphological features across thousands of images in one automated sweep. CellProfiler’s emphasis on reproducibility and scalability cemented its place as a cornerstone tool in large-scale phenotypic screening and drug discovery.
Suite2p
First released in 2017 by Marius Pachitariu and colleagues at HHMI Janelia, Suite2p (short for “Suite for 2-Photon imaging analysis”) aimed to streamline the workflow for calcium imaging experiments in neuroscience.
It offers automated solutions for image registration, region-of-interest detection, and neural signal extraction, all optimized for the massive data volumes characteristic of modern two-photon imaging studies. Suite2p’s ability to handle both speed and scale transformed it into a staple in many neurobiology labs investigating large neuronal populations.
Concrete scientific things you can do with these tools
Tool descriptions and history can be a bit abstract. For each tool, we’ll now outline a concrete biological use case to show the breadth of applications in scientific imaging.
Use Napari to visualize a 3D embryonic development dataset
Suppose you have a series of light-sheet microscopy images capturing an entire developing embryo over time (e.g., a zebrafish embryo). Using Napari, you can load these large, multi-channel ND datasets and scroll through different planes. You might apply a plugin to highlight fluorescently labeled cells (such as labeled nuclei or specific tissue markers) and annotate regions of interest in real time.
Because Napari supports GPU-accelerated rendering and has a flexible plugin ecosystem, you can navigate between timepoints, adjust brightness/contrast for different channels, and perform on-the-fly segmentation checks—significantly improving the speed and clarity of embryo staging experiments.
ImageJ for colocalization in immunofluorescent staining
In a cell biology experiment investigating whether two proteins (e.g., Protein X and Protein Y) co-localize within specific subcellular compartments, you can use ImageJ (or Fiji, its popular distribution) to measure the overlap of fluorescence signals.
By loading dual-color confocal images, applying background subtraction, and using plugins such as “Coloc 2,” you can generate metrics like Pearson’s correlation coefficient or Mander’s overlap coefficient. These measures provide quantitative evidence for co-localization, helping confirm whether Protein X truly interacts with Protein Y inside the same subcellular regions (like the nucleus or Golgi apparatus).
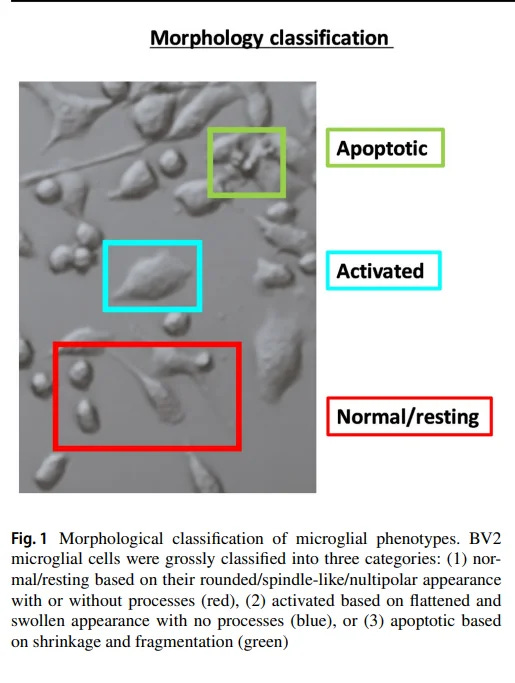
Cellpose to segment microglia in brain slices
If you’re studying how microglia (the resident immune cells in the brain) change their morphology in response to an injury or disease state, Cellpose can automatically identify and segment microglia in fluorescent images of stained brain tissue sections. After training or using the general pre-trained model, you can process hundreds of images, each containing various cell types.
Cellpose distinguishes cell boundaries even when they are partially overlapping or vary widely in shape. You can then quantify morphological features (like ramification index or cell body size) to assess how these immune cells respond under different experimental conditions.
CellProfiler for high-throughput cancer cell morphology screening
In a drug discovery project, you might test the effect of different small-molecule compounds on cancer cell morphology. By loading an entire 96- or 384-well plate worth of microscopy images into CellProfiler, you can design a pipeline to automatically segment nuclei and cell boundaries, measure dozens of features (such as cell size, shape, and intensity), and create a per-well summary of how each compound affects cell health or morphology.
Results can be exported in spreadsheet form for immediate statistical analysis, allowing you to rapidly identify promising hits or rule out toxic compounds early in your screening workflow.
Suite2p to map neural activity in a mouse visual cortex
In a two-photon calcium imaging experiment, you might record the activity of hundreds of neurons in the mouse visual cortex while showing various visual stimuli (like drifting gratings or natural scenes). With Suite2p, you can automatically correct for motion in the recorded images, identify the regions of interest (ROIs) corresponding to individual neurons, and extract the calcium signals that reflect neural firing.
By correlating these signals with the type of visual stimulus, you can pinpoint which neurons respond to certain orientations or patterns—a critical step in deciphering the cortical circuitry underlying visual processing.
Data infrastructure improves practical use of these tools
While scientists have been using these open-source tools on local computers for years, there are benefits to hosted data infrastructure that emerge with scale of imaging data, team size and project complexity.
The principles we discuss are not complex but have powerful effects on research efficiency:
Centralization of different tools
Libraries of containerized tool environments
Moving compute to data
Centralizing tools in a single place
This is just more efficient. Teammates no longer have to repeat setup steps on their own computers. They can just open a shared environment to access the latest templates or run new experiments. Unified systems for containerized tools, especially when colocated with experimental data, simplify the day to day of R&D.
Container “template” libraries prevent wasted effort on deployment
Rather than spending hours, or days, fiddling with drivers, conflicting dependencies and environment variables, container libraries provide “known-good” container templates from a shared repository, each with everything needed (e.g., Python libraries, GPU drivers, system packages) to run a particular imaging or analysis tool.
This solves for mundane work running and deploying these tools duplicated across hundreds of otherwise useful scientific minds.
These templates are also editable. They are just containers with complete access to source code and libraries. Changes get tracked, ensuring reproducibility—every lab member knows exactly which version of a tool or library was used.
The net result: no repeated wheel-reinvention for environment setup. Researchers jump straight into collecting data, analyzing results, and iterating on experiments.
Resize, duplicate and move tool compute to data
Latch has developed a portable computing environment that can be detached and moved to arbitrary machines. To do this, the system dumps a computer’s state—including libraries, code, environment configurations, and data—into a persistent storage layer. When you need more compute, you spin up a new container (or “Pod”) close to where your data resides. That container reattaches the same mount (using OverlayFS), eliminating bulky transfers or repeated uploads.
If the job outgrows your CPU or GPU allocation, you can spin down the smaller container and spin up a bigger one using the same mounted storage and environment. This agility is particularly helpful when analyzing gigabytes or terabytes of microscopy images, where running out of resources mid-analysis can be a major bottleneck.
By bringing code to imaging data instead of the other way around, labs save on network egress costs and reduce latency, allowing researchers to focus on scientific insights rather than data logistics. With image data generation growing rapidly, labs will be forced to adopt this analysis structure eventually
To conclude
The five open-source projects highlighted here - Napari, ImageJ, Cellpose, CellProfiler, Suite2p - allude to diverse and interesting use cases for imaging in biology. While useful on their own, hosting them together in a modern data infrastructure provides benefits for R&D efficiency.
These tools are available on Latch, a programmable data infrastructure built for biotech teams, as a library of Pod templates. Researchers can spin up, share, resize or duplicate these modular tools to analyze their data.