You can now run single-cell experiments in a tube with 4.5 hours of hands-on time
A new and accessible single-cell sequencing technology and its impact on progress in biotech
What happens when single-cell sequencing becomes cheaper, faster to process and easier? More scientists can access it, existing applications become more powerful and new types of experiments become possible.
While one of the most exciting technologies of the past decade, single-cell has been largely relegated to those with larger budgets and extensive lab expertise - academic consortiums, private institutes, well-funded biotechs.
Existing technologies require additional equipment, expensive reagent costs or suffer from multiple days of tedious lab workflows. They also require sequencing a large number of cells but only from a handful of samples, limiting experimental design to a specific type of low sample, replicate-free observational studies (eg. the beautiful tissue atlases we are all familiar with). The consequence? Many labs and companies still don’t use single-cell sequencing even though their research would greatly benefit. The ones that do have yet to adopt it for large scale perturbation studies, drug screens or population-scale variation studies useful for engineering molecules, developing drugs or understanding patient populations.
While there has been much development in new methods over the past few years, CS Genetics has developed a clever technique that allows scientists to index and prep cells entirely in-solution, using standard plastic labware, with 4.5 hours of hands-on time.
Here we dig into:
The technology
The data
What’s now possible
How accessible analysis integrates with SimpleCell
How does it work?
CS Genetics develops and sells kits that allow scientists to run cell to sequencer workflows in 0.2 ml tubes, creating an easier workflow and making true sample scaling possible. The technology is called SimpleCell and has two main components: a reagent with beads and a viscous buffer.
The first component is a reagent called CPair. It consists of beads coated with molecules that stick to cells and indexing oligos that bind to transcripts. Importantly, they are about the same size as most mammalian cells. When introduced to cells in precise ratios, the geometry of the beads favor 1:1 binding events with cells. This is the simple insight that allows for precise single cell indexing without other types of separation, like emulsion - leveraging the physical properties and stoichiometry of cells and beads are sufficient with careful engineering.
The second component is a viscous buffer called Kinetic Confinement Buffer that prevents the cell-bead pairs from moving around. After the buffer is added, the cells are lysed and transcripts bind with the indexed oligos from the beads. There are two properties of the mixture that are important - (1) transcripts from the cells remain in high concentration close to the bead and (2) the cell bead pairs remain separated from one another.
At this point, the sample tube contains indexed transcripts, uniquely tagged by the individual cell they came from. Creating cDNA and amplifying the fragments into sequencing-ready libraries can proceed in the same tube. The whole process takes 8 hours, of which 4.5 hours requires hands-on time.
In addition to making the workflow easy and cheap, a truly exciting aspect of this workflow is the sample scalability. Whether running 1 sample or 96 samples, both the economics and workflow are uniquely scalable. Want to sequence 96 samples? Use a 96-well plate instead of a strip tube to process your experiment.
How do we know it works?
When new techniques come online, it's important to check if they work. The simplicity and lower cost matters little if we are unable to accurately measure the genes of each cell at parity with what exists.
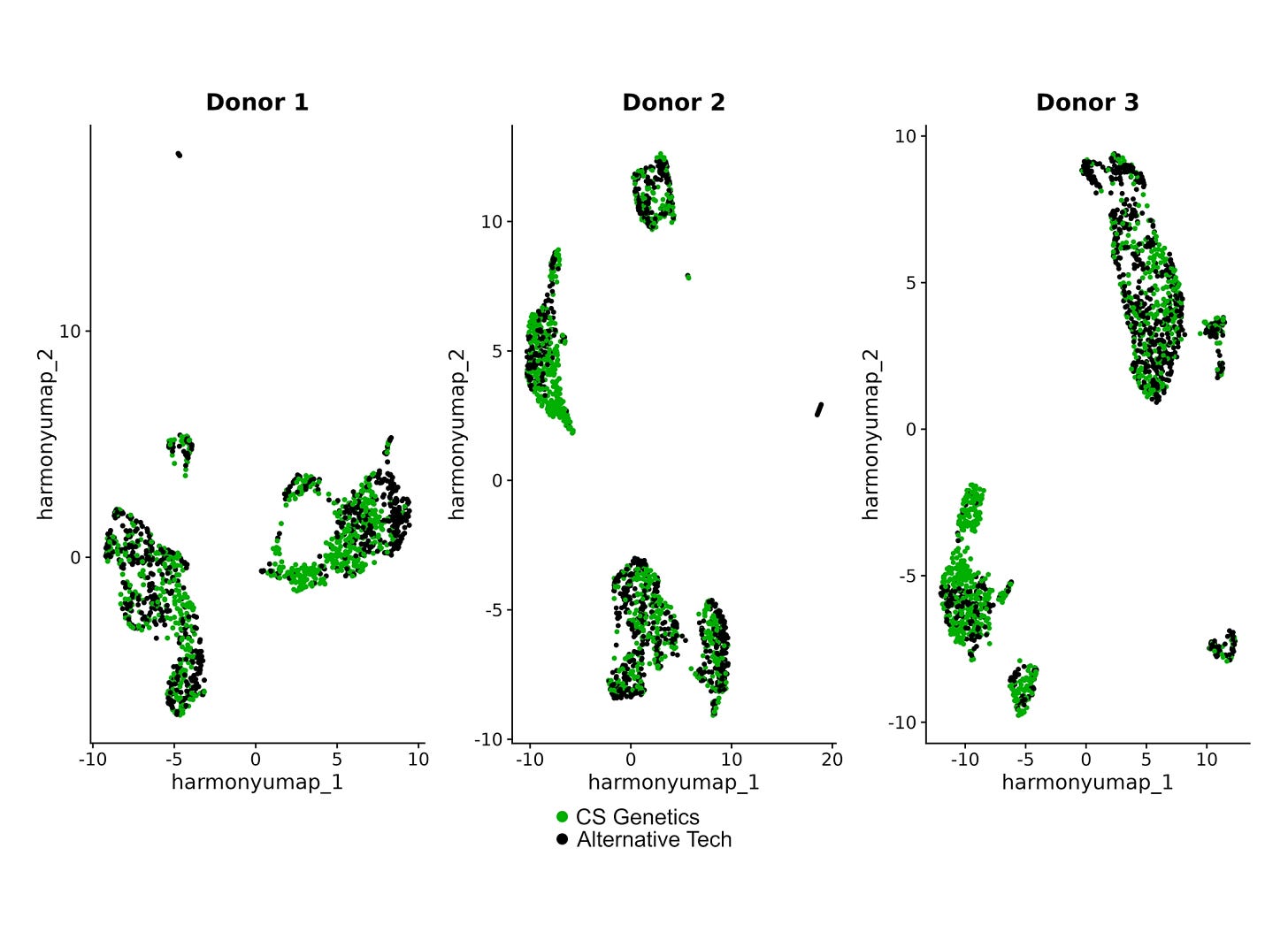
For single-cell, we can check this with a familiar gold-standard experiment - sequence a handful of PBMC samples and faithfully recover the different cell populations.
In Marifini et. al, 4 samples of 8.5K PBMC cells were sequenced after the SimpleCell workflow. With a sequencing depth of ~40K reads per cell, an average of 3.8K cells per sample were recovered, with a median transcript count of 2.6K and median gene count of 1.5K per cell.
We are able to recover the cell type populations in agreement with expected proportions.
What happens as you scale things up? In the same paper, 96 PBMC samples from 48 donors were prepared in parallel. Two samples from each donor were used to see if they produced similar results.
In this same batch, cell types identified from transcriptional data were compared against those identified with surface protein expression using data from a flow cytometry experiment.
What can we do with cost-effective and accessible Single-Cell Sequencing?
Packaging single-cell into budget-friendly, easy-to-use kits has exciting implications for new types of experiments. Immediately, the familiar class of data-rich atlasing studies become accessible to labs and fledgling biotechs across the world. Why do certain T-cells stick around in joint tissue and display autoimmune behavior in rheumatoid arthritis? Why do certain patients respond to immunotherapy in hepatocellular carcinoma and others do not? More studies, more data, more compounding knowledge.
But far more happens when you improve powerful tools. The research community doesn’t typically run single-cell experiments for repetitive, multi-sample experiments, but that will start to change.
Drug screens across banks of cell lines, perturbation studies and functional assays are the workhorse experiments of biotech, especially in iterative loops used to engineer libraries of molecules. Replacing or supplementing bulk sequencing and biochemical assays with high resolution single cell sequencing data at different legs of the drug development process will lead to unexpected discoveries (new targets, mechanisms, drug candidates), higher quality translational data and more repeatable campaigns with less follow-on experiments.
In clinical research, large population-scale variational studies to identify treatable patient populations and understand drug response are rare and undersized. We still use gene panels and bulk RNA sequencing to diagnose patients or indicate targeted treatment. We still use flow cytometry to screen immune cells for CAR-T.
Why accessible analysis is an important piece
SimpleCell is powerful because it puts experimental control into the hands of researchers. It instills confidence in their ability to understand and execute each step of the end-to-end workflow on their own. With the volume and complexity of single-cell sequencing data, analysis has been a large and difficult component of this workflow, standing in the way of fully understanding the outcomes of an experiment.
Here, bench scientist-friendly analysis portals complement the accessibility of SimpleCell kits. CS Genetics has partnered with Latch to provide access to bioinformatics workflows and graphical “notebooks” for secondary analysis. They bundle platform access with their kits to help biologists quickly get to clustering, cell-typing and visualization of gene expression and cell types.
Learn more about SimpleCell
If you would like to know more about CS Genetics’ SimpleCell technology and its workflow, please visit csgenetics.com or send an email to info@csgenetics.com.
–
Thank you to members of the CS Genetics team, Brian Steffy and Ben Hume, and Alfredo Andere for their thoughtful comments.