SpatialBench: real world tasks for spatial agents
98 dataset/eval packs across 4 spatial technologies // test harness, open formats and examples available on GitHub // eval banks as structured documentation of tacit knowledge
Agents are starting to work for spatial data analysis, but it’s often unclear how to measure their performance on real world tasks or rigorously compare different systems. Introducing SpatialBench, a suite of 98 dataset/eval packs constructed in collaboration with spatial vendors and scientists on tasks like cell typing, cell segmentation and spatially aware differential expression.
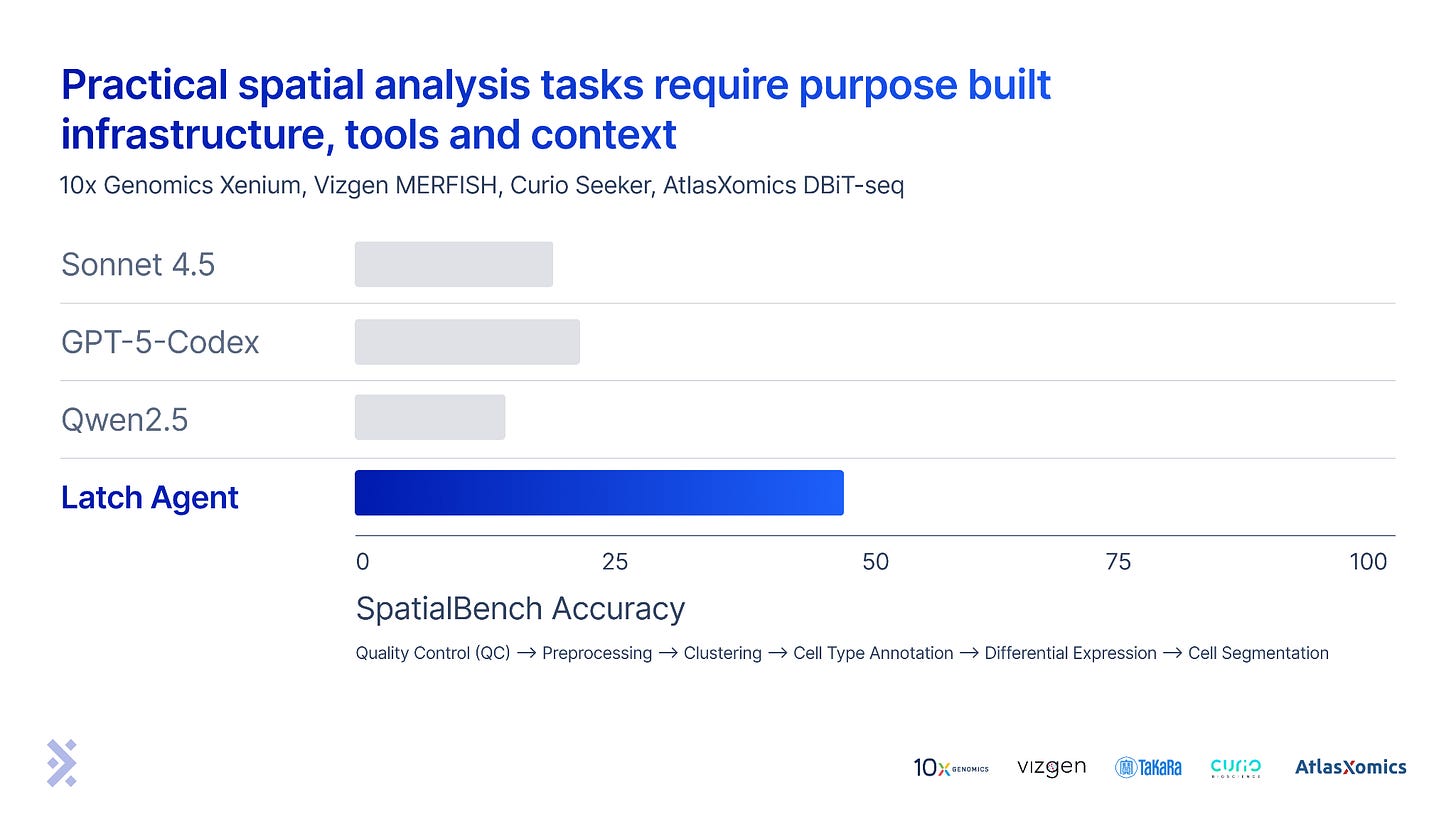
High scores on SpatialBench represent expert and industrially relevant analysis ability across four different spatial technologies, tissue types and diseases. We benchmarked our spatial agent against Sonnet 4.5, GPT-5-Codex and Qwen 2.5 Coder in a minimal harness (a slightly adapted version of the Bash only SWE-bench harness). These early results show considerable engineering is needed to extend computer use agents to biology. There is a lot of work remaining to saturate these benchmarks.
We’re also releasing a batched test harness, eval spec and a representative subset of tasks/datasets on GitHub. While we built this tool to measure and improve our own product, we hope others find the open resources useful.
SpatialBench evals represent practical analysis tasks
This benchmark is focused on real world analysis work and each task was constructed actual chunk of work from our customers and their scientists.
Each eval is a JSON object conforming to a standard structure: a task description, data node and grader configuration. This information is injected into a system prompt and the agent performs work in a sandbox with access to data. A grader then evaluates the answer.
Here we demonstrate some concrete examples to build some intuition for what the benchmark is actually measuring. The specs for these evals are available in the repo.
Normalization
Feature Selection
Dimensionality Reduction
Clustering
Differential Gene Expression
Domain Detection
Results demonstrate need for biology specific agent engineering
While the benchmark does not represent an apples-to-apples comparison of systems, eg. a ~100 line mini-swe-agent harness wrapping Sonnet is significantly less complex than our system, the purpose of the exercise was to show what worked for coding will not necessarily work for biology. We will be improving the harness and expanding the eval bank over the coming weeks.
Putting a model in a simple loop with access to a shell is sufficient to saturate SWE-bench, but agents for spatial biology likely need purpose built infrastructure, tools and context for useful work. More on the details here.
Evals encode tacit knowledge of frontier research biology
The frontier of research biology - practical knowledge about details of different diseases and tissues - is distributed across hundreds of labs, in the minds of PIs and researchers. By working with these labs to interpret spatial data from different tech, we can encode their knowledge and preferences into evals used to train and benchmark agentic systems. These eval banks will become valuable, structured documentation about the procedural details of research as they grow.
Add your spatial technology to our benchmarks
We are expanding SpatialBench to include all relevant machine and kit types. Reach out to our team to include your technology in our next release.